Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_XENOBIOTIC_METABOLISM |
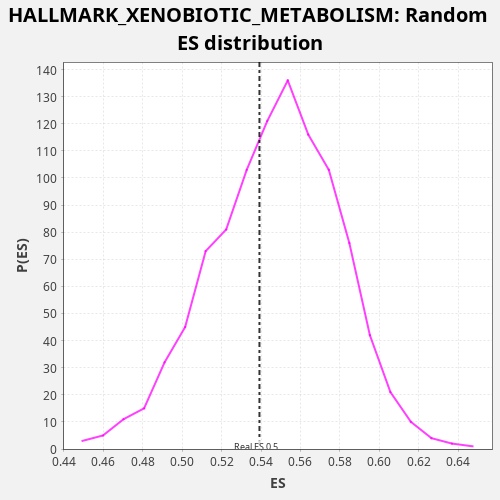
| Enrichment Score (ES) | 0.53921074 |
| Normalized Enrichment Score (NES) | 0.9856155 |
| Nominal p-value | 0.618 |
| FDR q-value | 0.80391806 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | CYP26A1 | 64 | 2.936 | 0.0405 | Yes |
| 2 | GAD1 | 165 | 2.483 | 0.0721 | Yes |
| 3 | FMO3 | 176 | 2.458 | 0.1084 | Yes |
| 4 | ETFDH | 199 | 2.396 | 0.1431 | Yes |
| 5 | TDO2 | 377 | 2.061 | 0.1641 | Yes |
| 6 | LONP1 | 528 | 1.897 | 0.1842 | Yes |
| 7 | ELOVL5 | 635 | 1.791 | 0.2051 | Yes |
| 8 | MTHFD1 | 769 | 1.677 | 0.2228 | Yes |
| 9 | SLC35B1 | 781 | 1.670 | 0.2472 | Yes |
| 10 | DHRS1 | 811 | 1.632 | 0.2701 | Yes |
| 11 | MT2A | 882 | 1.575 | 0.2897 | Yes |
| 12 | SLC46A3 | 936 | 1.539 | 0.3099 | Yes |
| 13 | PTGDS | 1436 | 1.237 | 0.3004 | Yes |
| 14 | PGD | 1639 | 1.121 | 0.3059 | Yes |
| 15 | GNMT | 1643 | 1.120 | 0.3225 | Yes |
| 16 | ALAS1 | 1677 | 1.108 | 0.3373 | Yes |
| 17 | PDLIM5 | 1754 | 1.073 | 0.3491 | Yes |
| 18 | ABCC3 | 1813 | 1.048 | 0.3616 | Yes |
| 19 | AP4B1 | 1816 | 1.046 | 0.3772 | Yes |
| 20 | MAN1A1 | 1886 | 1.013 | 0.3885 | Yes |
| 21 | FETUB | 1902 | 1.005 | 0.4027 | Yes |
| 22 | XDH | 1949 | 0.983 | 0.4149 | Yes |
| 23 | ACOX2 | 1998 | 0.964 | 0.4267 | Yes |
| 24 | ACP2 | 2096 | 0.924 | 0.4351 | Yes |
| 25 | BCAR1 | 2102 | 0.922 | 0.4486 | Yes |
| 26 | SLC35D1 | 2213 | 0.875 | 0.4556 | Yes |
| 27 | SMOX | 2251 | 0.859 | 0.4664 | Yes |
| 28 | KYNU | 2338 | 0.826 | 0.4740 | Yes |
| 29 | ADH7 | 2736 | 0.679 | 0.4619 | Yes |
| 30 | SSR3 | 2821 | 0.656 | 0.4670 | Yes |
| 31 | PAPSS2 | 2909 | 0.635 | 0.4716 | Yes |
| 32 | GART | 2944 | 0.625 | 0.4791 | Yes |
| 33 | IGFBP1 | 2977 | 0.618 | 0.4866 | Yes |
| 34 | NQO1 | 3131 | 0.580 | 0.4867 | Yes |
| 35 | PMM1 | 3178 | 0.571 | 0.4927 | Yes |
| 36 | CAT | 3323 | 0.539 | 0.4927 | Yes |
| 37 | FMO1 | 3326 | 0.538 | 0.5006 | Yes |
| 38 | ADH5 | 3633 | 0.479 | 0.4906 | Yes |
| 39 | MCCC2 | 3686 | 0.467 | 0.4947 | Yes |
| 40 | CYP2S1 | 3768 | 0.454 | 0.4970 | Yes |
| 41 | F11 | 3864 | 0.440 | 0.4982 | Yes |
| 42 | CYP27A1 | 3936 | 0.428 | 0.5007 | Yes |
| 43 | GCNT2 | 3971 | 0.424 | 0.5051 | Yes |
| 44 | CYP17A1 | 3983 | 0.422 | 0.5108 | Yes |
| 45 | CES1 | 4057 | 0.413 | 0.5129 | Yes |
| 46 | PDK4 | 4062 | 0.412 | 0.5189 | Yes |
| 47 | VTN | 4153 | 0.396 | 0.5198 | Yes |
| 48 | UPP1 | 4156 | 0.396 | 0.5256 | Yes |
| 49 | ARG2 | 4167 | 0.395 | 0.5310 | Yes |
| 50 | FBP1 | 4237 | 0.386 | 0.5329 | Yes |
| 51 | ITIH4 | 4281 | 0.380 | 0.5362 | Yes |
| 52 | GCKR | 4328 | 0.374 | 0.5392 | Yes |
| 53 | DDC | 4588 | 0.346 | 0.5299 | No |
| 54 | SLC12A4 | 4703 | 0.334 | 0.5285 | No |
| 55 | ABCC2 | 4788 | 0.326 | 0.5287 | No |
| 56 | DHRS7 | 4961 | 0.309 | 0.5236 | No |
| 57 | TTPA | 5012 | 0.305 | 0.5254 | No |
| 58 | CASP6 | 5043 | 0.302 | 0.5283 | No |
| 59 | GSS | 5077 | 0.299 | 0.5309 | No |
| 60 | ASL | 5237 | 0.284 | 0.5262 | No |
| 61 | HSD17B2 | 5293 | 0.280 | 0.5273 | No |
| 62 | ANGPTL3 | 5632 | 0.254 | 0.5122 | No |
| 63 | ENTPD5 | 5643 | 0.253 | 0.5154 | No |
| 64 | AKR1C3 | 5851 | 0.240 | 0.5074 | No |
| 65 | PTGR1 | 5946 | 0.234 | 0.5056 | No |
| 66 | FAH | 6044 | 0.229 | 0.5036 | No |
| 67 | ARG1 | 6092 | 0.227 | 0.5043 | No |
| 68 | ALDH2 | 6132 | 0.224 | 0.5055 | No |
| 69 | CNDP2 | 6153 | 0.223 | 0.5077 | No |
| 70 | ACP1 | 6198 | 0.221 | 0.5086 | No |
| 71 | ALDH3A1 | 6462 | 0.207 | 0.4969 | No |
| 72 | VNN1 | 6491 | 0.206 | 0.4985 | No |
| 73 | BPHL | 6589 | 0.201 | 0.4960 | No |
| 74 | CD36 | 6622 | 0.199 | 0.4972 | No |
| 75 | HRG | 6671 | 0.197 | 0.4975 | No |
| 76 | UPB1 | 6798 | 0.191 | 0.4933 | No |
| 77 | IDH1 | 6803 | 0.191 | 0.4959 | No |
| 78 | BLVRB | 6901 | 0.187 | 0.4932 | No |
| 79 | EPHA2 | 6958 | 0.185 | 0.4929 | No |
| 80 | SLC22A1 | 6998 | 0.182 | 0.4934 | No |
| 81 | RETSAT | 7043 | 0.181 | 0.4937 | No |
| 82 | CFB | 7136 | 0.177 | 0.4912 | No |
| 83 | SAR1B | 7268 | 0.172 | 0.4864 | No |
| 84 | HGFAC | 7489 | 0.164 | 0.4765 | No |
| 85 | ABCD2 | 7503 | 0.164 | 0.4782 | No |
| 86 | SLC6A12 | 7504 | 0.164 | 0.4807 | No |
| 87 | PSMB10 | 7588 | 0.161 | 0.4784 | No |
| 88 | KARS1 | 7597 | 0.161 | 0.4804 | No |
| 89 | JUP | 7605 | 0.160 | 0.4824 | No |
| 90 | AOX1 | 7664 | 0.158 | 0.4815 | No |
| 91 | PC | 7802 | 0.154 | 0.4761 | No |
| 92 | ACOX3 | 7878 | 0.152 | 0.4742 | No |
| 93 | TMBIM6 | 8018 | 0.147 | 0.4686 | No |
| 94 | IGFBP4 | 8061 | 0.145 | 0.4684 | No |
| 95 | HMOX1 | 8162 | 0.142 | 0.4649 | No |
| 96 | TPST1 | 8187 | 0.141 | 0.4657 | No |
| 97 | RBP4 | 8324 | 0.137 | 0.4601 | No |
| 98 | FBLN1 | 8430 | 0.134 | 0.4562 | No |
| 99 | PLG | 8459 | 0.133 | 0.4567 | No |
| 100 | UGDH | 8465 | 0.133 | 0.4584 | No |
| 101 | TMEM97 | 8515 | 0.132 | 0.4576 | No |
| 102 | LPIN2 | 8610 | 0.129 | 0.4543 | No |
| 103 | ACOX1 | 8727 | 0.126 | 0.4496 | No |
| 104 | CRP | 8983 | 0.119 | 0.4371 | No |
| 105 | TAT | 9032 | 0.119 | 0.4362 | No |
| 106 | HACL1 | 9054 | 0.118 | 0.4368 | No |
| 107 | TMEM176B | 9142 | 0.115 | 0.4336 | No |
| 108 | DHPS | 9193 | 0.114 | 0.4325 | No |
| 109 | TKFC | 9402 | 0.109 | 0.4225 | No |
| 110 | SHMT2 | 9422 | 0.109 | 0.4231 | No |
| 111 | CYP4F2 | 9429 | 0.109 | 0.4244 | No |
| 112 | ADH1C | 9522 | 0.107 | 0.4208 | No |
| 113 | MPP2 | 9556 | 0.106 | 0.4205 | No |
| 114 | AQP9 | 9712 | 0.103 | 0.4134 | No |
| 115 | ITIH1 | 9820 | 0.100 | 0.4089 | No |
| 116 | CYP2J2 | 9884 | 0.099 | 0.4068 | No |
| 117 | AKR1C2 | 9893 | 0.099 | 0.4078 | No |
| 118 | AHCY | 9972 | 0.097 | 0.4049 | No |
| 119 | LCAT | 10074 | 0.096 | 0.4007 | No |
| 120 | DCXR | 10118 | 0.095 | 0.3997 | No |
| 121 | ALDH9A1 | 10273 | 0.092 | 0.3924 | No |
| 122 | TYR | 10299 | 0.092 | 0.3924 | No |
| 123 | PYCR1 | 10380 | 0.090 | 0.3893 | No |
| 124 | GSTO1 | 10562 | 0.087 | 0.3804 | No |
| 125 | GABARAPL1 | 10601 | 0.086 | 0.3796 | No |
| 126 | POR | 10648 | 0.086 | 0.3783 | No |
| 127 | F10 | 10693 | 0.085 | 0.3771 | No |
| 128 | CDA | 10857 | 0.082 | 0.3692 | No |
| 129 | GCLC | 10980 | 0.080 | 0.3635 | No |
| 130 | ENPEP | 11148 | 0.077 | 0.3553 | No |
| 131 | DDAH2 | 11198 | 0.076 | 0.3537 | No |
| 132 | CDO1 | 11212 | 0.076 | 0.3541 | No |
| 133 | PTS | 11284 | 0.075 | 0.3512 | No |
| 134 | NDRG2 | 11578 | 0.070 | 0.3358 | No |
| 135 | CYP1A1 | 11706 | 0.068 | 0.3297 | No |
| 136 | ABHD6 | 11744 | 0.068 | 0.3287 | No |
| 137 | SERPINA6 | 11797 | 0.067 | 0.3268 | No |
| 138 | PROS1 | 11844 | 0.066 | 0.3252 | No |
| 139 | REG1A | 11940 | 0.065 | 0.3208 | No |
| 140 | CYFIP2 | 11948 | 0.064 | 0.3214 | No |
| 141 | ACO2 | 11958 | 0.064 | 0.3218 | No |
| 142 | CSAD | 12147 | 0.062 | 0.3122 | No |
| 143 | CYP2C18 | 12151 | 0.062 | 0.3130 | No |
| 144 | GSTA3 | 12276 | 0.060 | 0.3069 | No |
| 145 | CA2 | 12332 | 0.060 | 0.3047 | No |
| 146 | FABP1 | 12730 | 0.054 | 0.2832 | No |
| 147 | LEAP2 | 12733 | 0.054 | 0.2839 | No |
| 148 | MARCHF6 | 12742 | 0.054 | 0.2843 | No |
| 149 | NFS1 | 12830 | 0.053 | 0.2802 | No |
| 150 | CYP1A2 | 13103 | 0.049 | 0.2657 | No |
| 151 | MBL2 | 13168 | 0.048 | 0.2628 | No |
| 152 | IL1R1 | 13193 | 0.048 | 0.2622 | No |
| 153 | SLC6A6 | 13244 | 0.047 | 0.2601 | No |
| 154 | SLC1A5 | 13265 | 0.047 | 0.2596 | No |
| 155 | BCAT1 | 13556 | 0.043 | 0.2440 | No |
| 156 | COMT | 13602 | 0.042 | 0.2421 | No |
| 157 | CYB5A | 13609 | 0.042 | 0.2424 | No |
| 158 | ATOH8 | 13843 | 0.039 | 0.2299 | No |
| 159 | NPC1 | 13967 | 0.037 | 0.2235 | No |
| 160 | CBR1 | 14017 | 0.036 | 0.2213 | No |
| 161 | EPHX1 | 14047 | 0.035 | 0.2202 | No |
| 162 | PGRMC1 | 14526 | 0.011 | 0.1936 | No |
| 163 | CROT | 14549 | -0.000 | 0.1923 | No |
| 164 | IGF1 | 14601 | -0.000 | 0.1895 | No |
| 165 | TNFRSF1A | 14698 | -0.000 | 0.1841 | No |
| 166 | ESR1 | 14862 | -0.000 | 0.1749 | No |
| 167 | TGFB2 | 14869 | -0.000 | 0.1746 | No |
| 168 | DDT | 14891 | -0.000 | 0.1734 | No |
| 169 | HNF4A | 14935 | -0.000 | 0.1710 | No |
| 170 | GSR | 15048 | -0.000 | 0.1647 | No |
| 171 | PTGES3 | 15207 | -0.000 | 0.1558 | No |
| 172 | PPARD | 15235 | -0.000 | 0.1543 | No |
| 173 | ID2 | 15330 | -0.000 | 0.1490 | No |
| 174 | HSD11B1 | 15371 | -0.000 | 0.1468 | No |
| 175 | ARPP19 | 15643 | -0.000 | 0.1316 | No |
| 176 | APOE | 15671 | -0.000 | 0.1301 | No |
| 177 | CYP2E1 | 15682 | -0.000 | 0.1295 | No |
| 178 | NINJ1 | 15718 | -0.000 | 0.1275 | No |
| 179 | GCH1 | 15732 | -0.000 | 0.1268 | No |
| 180 | NMT1 | 15848 | -0.000 | 0.1204 | No |
| 181 | IRF8 | 15981 | -0.000 | 0.1130 | No |
| 182 | HES6 | 16060 | -0.000 | 0.1086 | No |
| 183 | PTGES | 16145 | -0.000 | 0.1039 | No |
| 184 | ETS2 | 16305 | -0.000 | 0.0949 | No |
| 185 | PINK1 | 16338 | -0.000 | 0.0931 | No |
| 186 | HPRT1 | 16577 | -0.000 | 0.0798 | No |
| 187 | ACSM1 | 16619 | -0.000 | 0.0775 | No |
| 188 | GSTM4 | 16709 | -0.000 | 0.0725 | No |
| 189 | ATP2A2 | 16936 | -0.000 | 0.0598 | No |
| 190 | MAOA | 17408 | -0.000 | 0.0333 | No |
| 191 | SERTAD1 | 17479 | -0.000 | 0.0294 | No |