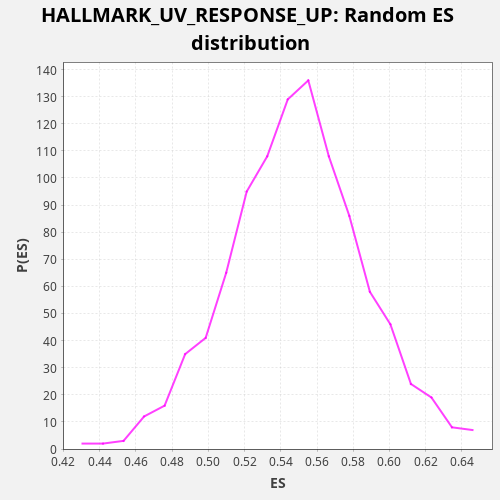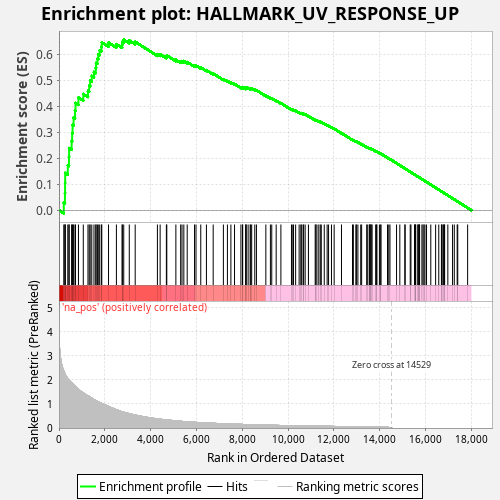
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_UV_RESPONSE_UP |
| Enrichment Score (ES) | 0.6566669 |
| Normalized Enrichment Score (NES) | 1.1973705 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.14500004 |
| FWER p-Value | 0.36 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | CCNE1 | 209 | 2.370 | 0.0297 | Yes |
| 2 | SLC25A4 | 260 | 2.250 | 0.0661 | Yes |
| 3 | POLR2H | 266 | 2.237 | 0.1049 | Yes |
| 4 | NFKBIA | 270 | 2.233 | 0.1437 | Yes |
| 5 | MMP14 | 384 | 2.049 | 0.1731 | Yes |
| 6 | C4BPB | 435 | 1.992 | 0.2051 | Yes |
| 7 | CCND3 | 442 | 1.984 | 0.2394 | Yes |
| 8 | FURIN | 551 | 1.878 | 0.2661 | Yes |
| 9 | MAPK8IP2 | 574 | 1.854 | 0.2973 | Yes |
| 10 | LHX2 | 594 | 1.837 | 0.3283 | Yes |
| 11 | CASP3 | 637 | 1.789 | 0.3571 | Yes |
| 12 | CREG1 | 699 | 1.737 | 0.3840 | Yes |
| 13 | IL6ST | 721 | 1.721 | 0.4129 | Yes |
| 14 | TUBA4A | 849 | 1.603 | 0.4338 | Yes |
| 15 | TGFBRAP1 | 1060 | 1.459 | 0.4475 | Yes |
| 16 | CHKA | 1265 | 1.338 | 0.4594 | Yes |
| 17 | RAB27A | 1318 | 1.311 | 0.4794 | Yes |
| 18 | CHRNA5 | 1361 | 1.279 | 0.4994 | Yes |
| 19 | DDX21 | 1431 | 1.242 | 0.5172 | Yes |
| 20 | CTSV | 1527 | 1.183 | 0.5325 | Yes |
| 21 | RRAD | 1606 | 1.139 | 0.5480 | Yes |
| 22 | DNAJB1 | 1619 | 1.130 | 0.5671 | Yes |
| 23 | ALAS1 | 1677 | 1.108 | 0.5832 | Yes |
| 24 | CYB5R1 | 1714 | 1.093 | 0.6003 | Yes |
| 25 | CXCL2 | 1775 | 1.064 | 0.6155 | Yes |
| 26 | POLE3 | 1848 | 1.030 | 0.6294 | Yes |
| 27 | CDKN2B | 1868 | 1.019 | 0.6462 | Yes |
| 28 | TYRO3 | 2162 | 0.897 | 0.6454 | Yes |
| 29 | ATP6V1C1 | 2506 | 0.763 | 0.6395 | Yes |
| 30 | HYAL2 | 2755 | 0.674 | 0.6374 | Yes |
| 31 | ABCB1 | 2765 | 0.671 | 0.6486 | Yes |
| 32 | BTG3 | 2826 | 0.655 | 0.6567 | Yes |
| 33 | TARS1 | 3068 | 0.595 | 0.6535 | No |
| 34 | FMO1 | 3326 | 0.538 | 0.6485 | No |
| 35 | GPX3 | 4296 | 0.379 | 0.6009 | No |
| 36 | SULT1A1 | 4415 | 0.365 | 0.6006 | No |
| 37 | SPR | 4687 | 0.336 | 0.5913 | No |
| 38 | SQSTM1 | 4706 | 0.334 | 0.5962 | No |
| 39 | E2F5 | 5102 | 0.298 | 0.5792 | No |
| 40 | HNRNPU | 5311 | 0.279 | 0.5724 | No |
| 41 | TST | 5370 | 0.273 | 0.5740 | No |
| 42 | POLG2 | 5450 | 0.267 | 0.5742 | No |
| 43 | HSPA13 | 5597 | 0.257 | 0.5705 | No |
| 44 | PPIF | 5916 | 0.236 | 0.5568 | No |
| 45 | RFC4 | 5981 | 0.232 | 0.5573 | No |
| 46 | HLA-F | 6189 | 0.221 | 0.5495 | No |
| 47 | NR4A1 | 6436 | 0.209 | 0.5394 | No |
| 48 | SIGMAR1 | 6731 | 0.194 | 0.5263 | No |
| 49 | TAP1 | 7176 | 0.175 | 0.5045 | No |
| 50 | TACR3 | 7352 | 0.169 | 0.4976 | No |
| 51 | SLC6A12 | 7504 | 0.164 | 0.4920 | No |
| 52 | BAK1 | 7663 | 0.158 | 0.4860 | No |
| 53 | ASNS | 7942 | 0.149 | 0.4730 | No |
| 54 | PSMC3 | 8012 | 0.147 | 0.4717 | No |
| 55 | TMBIM6 | 8018 | 0.147 | 0.4740 | No |
| 56 | STK25 | 8151 | 0.142 | 0.4691 | No |
| 57 | HMOX1 | 8162 | 0.142 | 0.4710 | No |
| 58 | CDC34 | 8170 | 0.142 | 0.4731 | No |
| 59 | ICAM1 | 8256 | 0.139 | 0.4707 | No |
| 60 | ACAA1 | 8344 | 0.136 | 0.4682 | No |
| 61 | BID | 8369 | 0.135 | 0.4692 | No |
| 62 | STARD3 | 8412 | 0.134 | 0.4692 | No |
| 63 | PDLIM3 | 8547 | 0.131 | 0.4640 | No |
| 64 | PTPRD | 8619 | 0.129 | 0.4623 | No |
| 65 | BSG | 9030 | 0.119 | 0.4414 | No |
| 66 | UROD | 9229 | 0.113 | 0.4323 | No |
| 67 | EPCAM | 9288 | 0.112 | 0.4310 | No |
| 68 | IGFBP2 | 9481 | 0.108 | 0.4221 | No |
| 69 | PARP2 | 9687 | 0.103 | 0.4124 | No |
| 70 | TFRC | 10149 | 0.094 | 0.3882 | No |
| 71 | HTR7 | 10207 | 0.093 | 0.3867 | No |
| 72 | CLCN2 | 10239 | 0.093 | 0.3865 | No |
| 73 | MARK2 | 10331 | 0.091 | 0.3830 | No |
| 74 | SLC6A8 | 10483 | 0.089 | 0.3761 | No |
| 75 | RASGRP1 | 10560 | 0.087 | 0.3734 | No |
| 76 | BTG2 | 10578 | 0.087 | 0.3739 | No |
| 77 | WIZ | 10654 | 0.085 | 0.3712 | No |
| 78 | SELENOW | 10679 | 0.085 | 0.3714 | No |
| 79 | KCNH2 | 10746 | 0.084 | 0.3691 | No |
| 80 | GLS | 10889 | 0.081 | 0.3626 | No |
| 81 | AP2S1 | 11184 | 0.076 | 0.3475 | No |
| 82 | CDO1 | 11212 | 0.076 | 0.3473 | No |
| 83 | FKBP4 | 11260 | 0.075 | 0.3460 | No |
| 84 | ALDOA | 11342 | 0.074 | 0.3427 | No |
| 85 | ATP6V1F | 11407 | 0.073 | 0.3404 | No |
| 86 | PRKCD | 11450 | 0.072 | 0.3393 | No |
| 87 | CCK | 11580 | 0.070 | 0.3333 | No |
| 88 | CYP1A1 | 11706 | 0.068 | 0.3275 | No |
| 89 | DNAJA1 | 11768 | 0.067 | 0.3253 | No |
| 90 | CDK2 | 11895 | 0.065 | 0.3194 | No |
| 91 | ENO2 | 12012 | 0.064 | 0.3140 | No |
| 92 | CA2 | 12332 | 0.060 | 0.2971 | No |
| 93 | ATF3 | 12813 | 0.053 | 0.2712 | No |
| 94 | AQP3 | 12850 | 0.052 | 0.2701 | No |
| 95 | NPTXR | 12962 | 0.051 | 0.2647 | No |
| 96 | GAL | 12981 | 0.051 | 0.2646 | No |
| 97 | PPAT | 13048 | 0.050 | 0.2618 | No |
| 98 | RXRB | 13172 | 0.048 | 0.2557 | No |
| 99 | NPTX2 | 13212 | 0.048 | 0.2544 | No |
| 100 | RPN1 | 13432 | 0.045 | 0.2429 | No |
| 101 | FEN1 | 13478 | 0.044 | 0.2412 | No |
| 102 | CYB5B | 13549 | 0.043 | 0.2380 | No |
| 103 | AMD1 | 13570 | 0.042 | 0.2376 | No |
| 104 | NUP58 | 13580 | 0.042 | 0.2378 | No |
| 105 | GRPEL1 | 13615 | 0.042 | 0.2367 | No |
| 106 | SOD2 | 13630 | 0.042 | 0.2366 | No |
| 107 | EIF5 | 13669 | 0.041 | 0.2352 | No |
| 108 | BCL2L11 | 13825 | 0.039 | 0.2272 | No |
| 109 | CNP | 13857 | 0.039 | 0.2261 | No |
| 110 | MGAT1 | 13880 | 0.038 | 0.2256 | No |
| 111 | APOM | 13988 | 0.036 | 0.2202 | No |
| 112 | EPHX1 | 14047 | 0.035 | 0.2176 | No |
| 113 | ARRB2 | 14053 | 0.035 | 0.2179 | No |
| 114 | EIF2S3 | 14341 | 0.027 | 0.2023 | No |
| 115 | BTG1 | 14396 | 0.022 | 0.1997 | No |
| 116 | BMP2 | 14448 | 0.020 | 0.1972 | No |
| 117 | PRKACA | 14736 | -0.000 | 0.1811 | No |
| 118 | CDC5L | 14879 | -0.000 | 0.1731 | No |
| 119 | PDAP1 | 15099 | -0.000 | 0.1609 | No |
| 120 | YKT6 | 15111 | -0.000 | 0.1603 | No |
| 121 | PLCL1 | 15333 | -0.000 | 0.1479 | No |
| 122 | PRPF3 | 15364 | -0.000 | 0.1462 | No |
| 123 | AGO2 | 15520 | -0.000 | 0.1375 | No |
| 124 | KLHDC3 | 15547 | -0.000 | 0.1361 | No |
| 125 | IRF1 | 15558 | -0.000 | 0.1355 | No |
| 126 | FOSB | 15574 | -0.000 | 0.1347 | No |
| 127 | CDKN1C | 15664 | -0.000 | 0.1297 | No |
| 128 | OLFM1 | 15679 | -0.000 | 0.1289 | No |
| 129 | PPT1 | 15706 | -0.000 | 0.1274 | No |
| 130 | GCH1 | 15732 | -0.000 | 0.1260 | No |
| 131 | DLG4 | 15742 | -0.000 | 0.1255 | No |
| 132 | IL6 | 15843 | -0.000 | 0.1199 | No |
| 133 | GGH | 15891 | -0.000 | 0.1173 | No |
| 134 | COL2A1 | 15935 | -0.000 | 0.1149 | No |
| 135 | NTRK3 | 15976 | -0.000 | 0.1126 | No |
| 136 | RHOB | 16045 | -0.000 | 0.1088 | No |
| 137 | CEBPG | 16231 | -0.000 | 0.0985 | No |
| 138 | MSX1 | 16442 | -0.000 | 0.0867 | No |
| 139 | RET | 16579 | -0.000 | 0.0791 | No |
| 140 | STIP1 | 16695 | -0.000 | 0.0726 | No |
| 141 | SHOX2 | 16710 | -0.000 | 0.0719 | No |
| 142 | ONECUT1 | 16762 | -0.000 | 0.0690 | No |
| 143 | FOS | 16780 | -0.000 | 0.0681 | No |
| 144 | JUNB | 16825 | -0.000 | 0.0656 | No |
| 145 | NAT1 | 16835 | -0.000 | 0.0651 | No |
| 146 | CLTB | 16975 | -0.000 | 0.0573 | No |
| 147 | NKX2-5 | 17181 | -0.000 | 0.0458 | No |
| 148 | DGAT1 | 17257 | -0.000 | 0.0416 | No |
| 149 | H2AX | 17383 | -0.000 | 0.0346 | No |
| 150 | MAOA | 17408 | -0.000 | 0.0333 | No |
| 151 | LYN | 17839 | -0.000 | 0.0092 | No |