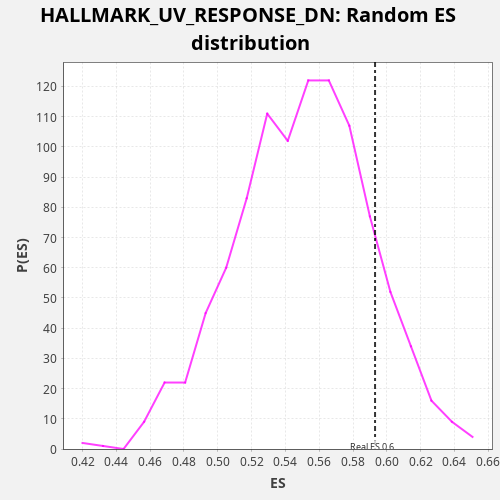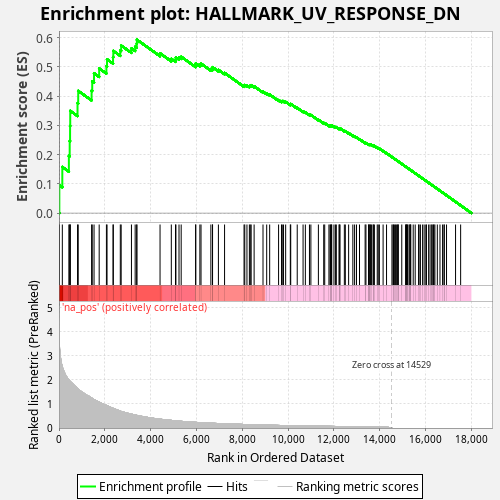
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_UV_RESPONSE_DN |
| Enrichment Score (ES) | 0.5930147 |
| Normalized Enrichment Score (NES) | 1.0778884 |
| Nominal p-value | 0.136 |
| FDR q-value | 0.47506693 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | MGLL | 15 | 3.569 | 0.0956 | Yes |
| 2 | PHF3 | 145 | 2.571 | 0.1578 | Yes |
| 3 | FZD2 | 431 | 1.996 | 0.1957 | Yes |
| 4 | F3 | 467 | 1.956 | 0.2466 | Yes |
| 5 | ACVR2A | 484 | 1.935 | 0.2979 | Yes |
| 6 | COL11A1 | 491 | 1.928 | 0.3497 | Yes |
| 7 | MET | 809 | 1.633 | 0.3760 | Yes |
| 8 | GCNT1 | 839 | 1.611 | 0.4179 | Yes |
| 9 | RASA2 | 1423 | 1.250 | 0.4190 | Yes |
| 10 | MIOS | 1451 | 1.227 | 0.4507 | Yes |
| 11 | IGF1R | 1530 | 1.180 | 0.4782 | Yes |
| 12 | PDLIM5 | 1754 | 1.073 | 0.4947 | Yes |
| 13 | AMPH | 2070 | 0.937 | 0.5023 | Yes |
| 14 | LDLR | 2097 | 0.924 | 0.5258 | Yes |
| 15 | ADORA2B | 2359 | 0.818 | 0.5333 | Yes |
| 16 | MAP2K5 | 2371 | 0.814 | 0.5547 | Yes |
| 17 | ATRN | 2676 | 0.700 | 0.5566 | Yes |
| 18 | PIAS3 | 2714 | 0.686 | 0.5730 | Yes |
| 19 | ITGB3 | 3163 | 0.573 | 0.5634 | Yes |
| 20 | BCKDHB | 3325 | 0.538 | 0.5689 | Yes |
| 21 | CDON | 3393 | 0.524 | 0.5793 | Yes |
| 22 | PTPN21 | 3401 | 0.522 | 0.5930 | Yes |
| 23 | VLDLR | 4411 | 0.366 | 0.5464 | No |
| 24 | SCAF8 | 4903 | 0.315 | 0.5274 | No |
| 25 | TGFBR2 | 5086 | 0.299 | 0.5253 | No |
| 26 | RGS4 | 5104 | 0.297 | 0.5324 | No |
| 27 | PRDM2 | 5239 | 0.284 | 0.5325 | No |
| 28 | APBB2 | 5334 | 0.276 | 0.5347 | No |
| 29 | PIK3R3 | 5967 | 0.233 | 0.5056 | No |
| 30 | SLC22A18 | 5968 | 0.233 | 0.5119 | No |
| 31 | VAV2 | 6150 | 0.224 | 0.5078 | No |
| 32 | ANXA4 | 6200 | 0.221 | 0.5111 | No |
| 33 | KCNMA1 | 6630 | 0.199 | 0.4924 | No |
| 34 | MT1E | 6702 | 0.195 | 0.4937 | No |
| 35 | TENT4A | 6703 | 0.195 | 0.4990 | No |
| 36 | IRS1 | 6959 | 0.185 | 0.4897 | No |
| 37 | TJP1 | 7228 | 0.173 | 0.4794 | No |
| 38 | BDNF | 8078 | 0.145 | 0.4358 | No |
| 39 | PDGFRB | 8110 | 0.144 | 0.4379 | No |
| 40 | TGFBR3 | 8205 | 0.141 | 0.4364 | No |
| 41 | ERBB2 | 8311 | 0.137 | 0.4343 | No |
| 42 | CACNA1A | 8350 | 0.136 | 0.4358 | No |
| 43 | ID1 | 8389 | 0.135 | 0.4373 | No |
| 44 | CAP2 | 8518 | 0.132 | 0.4337 | No |
| 45 | PLCB4 | 8906 | 0.121 | 0.4153 | No |
| 46 | DLC1 | 9067 | 0.117 | 0.4095 | No |
| 47 | MRPS31 | 9197 | 0.114 | 0.4054 | No |
| 48 | TOGARAM1 | 9586 | 0.105 | 0.3865 | No |
| 49 | SRI | 9713 | 0.103 | 0.3822 | No |
| 50 | ANXA2 | 9746 | 0.102 | 0.3832 | No |
| 51 | ICA1 | 9802 | 0.101 | 0.3828 | No |
| 52 | MGMT | 9895 | 0.099 | 0.3804 | No |
| 53 | MAGI2 | 10098 | 0.095 | 0.3716 | No |
| 54 | DDAH1 | 10116 | 0.095 | 0.3732 | No |
| 55 | DLG1 | 10400 | 0.090 | 0.3598 | No |
| 56 | ADD3 | 10655 | 0.085 | 0.3479 | No |
| 57 | COL1A1 | 10755 | 0.084 | 0.3446 | No |
| 58 | PRKCA | 10934 | 0.080 | 0.3368 | No |
| 59 | DMAC2L | 10996 | 0.079 | 0.3356 | No |
| 60 | ATP2B4 | 11324 | 0.074 | 0.3193 | No |
| 61 | SFMBT1 | 11556 | 0.071 | 0.3082 | No |
| 62 | CAV1 | 11600 | 0.070 | 0.3077 | No |
| 63 | ADGRL2 | 11784 | 0.067 | 0.2993 | No |
| 64 | NRP1 | 11833 | 0.066 | 0.2984 | No |
| 65 | AGGF1 | 11854 | 0.066 | 0.2991 | No |
| 66 | NR1D2 | 11899 | 0.065 | 0.2984 | No |
| 67 | MTA1 | 11905 | 0.065 | 0.2999 | No |
| 68 | NOTCH2 | 12003 | 0.064 | 0.2961 | No |
| 69 | FHL2 | 12068 | 0.063 | 0.2943 | No |
| 70 | INPP4B | 12107 | 0.062 | 0.2938 | No |
| 71 | COL1A2 | 12228 | 0.061 | 0.2887 | No |
| 72 | DAB2 | 12238 | 0.061 | 0.2899 | No |
| 73 | DYRK1A | 12269 | 0.060 | 0.2898 | No |
| 74 | GRK5 | 12457 | 0.058 | 0.2809 | No |
| 75 | ATP2C1 | 12500 | 0.057 | 0.2801 | No |
| 76 | LAMC1 | 12644 | 0.055 | 0.2736 | No |
| 77 | INSIG1 | 12828 | 0.053 | 0.2648 | No |
| 78 | PTGFR | 12905 | 0.052 | 0.2619 | No |
| 79 | FBLN5 | 12987 | 0.051 | 0.2587 | No |
| 80 | COL5A2 | 13119 | 0.049 | 0.2527 | No |
| 81 | NEK7 | 13364 | 0.046 | 0.2403 | No |
| 82 | COL3A1 | 13400 | 0.045 | 0.2396 | No |
| 83 | SIPA1L1 | 13513 | 0.043 | 0.2345 | No |
| 84 | GJA1 | 13523 | 0.043 | 0.2351 | No |
| 85 | PTEN | 13565 | 0.042 | 0.2340 | No |
| 86 | KIT | 13574 | 0.042 | 0.2347 | No |
| 87 | PIK3CD | 13604 | 0.042 | 0.2342 | No |
| 88 | WDR37 | 13646 | 0.042 | 0.2330 | No |
| 89 | PEX14 | 13716 | 0.041 | 0.2303 | No |
| 90 | PRKAR2B | 13722 | 0.041 | 0.2311 | No |
| 91 | SCN8A | 13769 | 0.040 | 0.2296 | No |
| 92 | NR3C1 | 13893 | 0.038 | 0.2237 | No |
| 93 | PPARG | 13937 | 0.037 | 0.2223 | No |
| 94 | DBP | 13987 | 0.036 | 0.2206 | No |
| 95 | DUSP1 | 14149 | 0.033 | 0.2124 | No |
| 96 | ABCC1 | 14299 | 0.029 | 0.2049 | No |
| 97 | TFPI | 14535 | -0.000 | 0.1917 | No |
| 98 | FYN | 14583 | -0.000 | 0.1891 | No |
| 99 | SNAI2 | 14604 | -0.000 | 0.1880 | No |
| 100 | CELF2 | 14652 | -0.000 | 0.1853 | No |
| 101 | CDK13 | 14686 | -0.000 | 0.1835 | No |
| 102 | ATP2B1 | 14728 | -0.000 | 0.1812 | No |
| 103 | SYNJ2 | 14773 | -0.000 | 0.1787 | No |
| 104 | YTHDC1 | 14804 | -0.000 | 0.1771 | No |
| 105 | ATRX | 14811 | -0.000 | 0.1767 | No |
| 106 | SMAD7 | 14964 | -0.000 | 0.1682 | No |
| 107 | BMPR1A | 15132 | -0.000 | 0.1589 | No |
| 108 | ZMIZ1 | 15146 | -0.000 | 0.1581 | No |
| 109 | PMP22 | 15171 | -0.000 | 0.1568 | No |
| 110 | NFKB1 | 15179 | -0.000 | 0.1564 | No |
| 111 | CDKN1B | 15217 | -0.000 | 0.1543 | No |
| 112 | MAPK14 | 15236 | -0.000 | 0.1533 | No |
| 113 | EFEMP1 | 15317 | -0.000 | 0.1488 | No |
| 114 | IGFBP5 | 15320 | -0.000 | 0.1487 | No |
| 115 | RND3 | 15336 | -0.000 | 0.1479 | No |
| 116 | AKT3 | 15358 | -0.000 | 0.1467 | No |
| 117 | SPOP | 15463 | -0.000 | 0.1409 | No |
| 118 | ATXN1 | 15550 | -0.000 | 0.1361 | No |
| 119 | ARHGEF9 | 15697 | -0.000 | 0.1279 | No |
| 120 | MAP1B | 15719 | -0.000 | 0.1267 | No |
| 121 | BHLHE40 | 15778 | -0.000 | 0.1235 | No |
| 122 | MYC | 15873 | -0.000 | 0.1182 | No |
| 123 | SLC7A1 | 15940 | -0.000 | 0.1145 | No |
| 124 | CCN1 | 16014 | -0.000 | 0.1104 | No |
| 125 | CDC42BPA | 16042 | -0.000 | 0.1089 | No |
| 126 | NFIB | 16137 | -0.000 | 0.1037 | No |
| 127 | SCHIP1 | 16199 | -0.000 | 0.1003 | No |
| 128 | MMP16 | 16270 | -0.000 | 0.0963 | No |
| 129 | RBPMS | 16293 | -0.000 | 0.0951 | No |
| 130 | RUNX1 | 16348 | -0.000 | 0.0921 | No |
| 131 | KALRN | 16370 | -0.000 | 0.0909 | No |
| 132 | PLPP3 | 16410 | -0.000 | 0.0887 | No |
| 133 | CITED2 | 16516 | -0.000 | 0.0828 | No |
| 134 | SMAD3 | 16632 | -0.000 | 0.0764 | No |
| 135 | SDC2 | 16745 | -0.000 | 0.0701 | No |
| 136 | HAS2 | 16809 | -0.000 | 0.0666 | No |
| 137 | PRKCE | 16820 | -0.000 | 0.0661 | No |
| 138 | PTPRM | 16918 | -0.000 | 0.0606 | No |
| 139 | RXRA | 17312 | -0.000 | 0.0386 | No |
| 140 | LPAR1 | 17534 | -0.000 | 0.0263 | No |