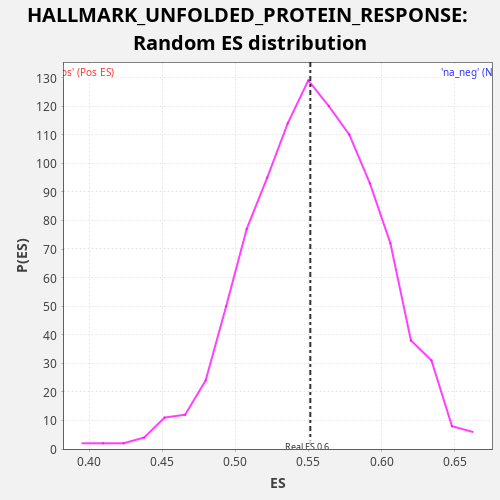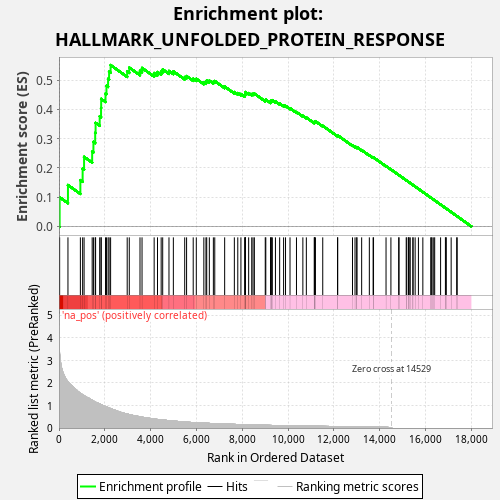
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.5512895 |
| Normalized Enrichment Score (NES) | 0.9952821 |
| Nominal p-value | 0.523 |
| FDR q-value | 0.8461241 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | YIF1A | 28 | 3.325 | 0.0996 | Yes |
| 2 | CNOT4 | 391 | 2.038 | 0.1414 | Yes |
| 3 | SDAD1 | 933 | 1.541 | 0.1581 | Yes |
| 4 | DDIT4 | 1032 | 1.475 | 0.1975 | Yes |
| 5 | PREB | 1095 | 1.436 | 0.2378 | Yes |
| 6 | DNAJA4 | 1448 | 1.231 | 0.2556 | Yes |
| 7 | EXOSC10 | 1503 | 1.196 | 0.2890 | Yes |
| 8 | EXOSC1 | 1580 | 1.157 | 0.3200 | Yes |
| 9 | HSP90B1 | 1598 | 1.145 | 0.3539 | Yes |
| 10 | EXOC2 | 1776 | 1.064 | 0.3764 | Yes |
| 11 | ATF6 | 1836 | 1.037 | 0.4046 | Yes |
| 12 | SRPRA | 1842 | 1.035 | 0.4358 | Yes |
| 13 | MTHFD2 | 2031 | 0.951 | 0.4543 | Yes |
| 14 | EIF4G1 | 2072 | 0.936 | 0.4805 | Yes |
| 15 | PDIA5 | 2143 | 0.908 | 0.5043 | Yes |
| 16 | TSPYL2 | 2187 | 0.885 | 0.5288 | Yes |
| 17 | KIF5B | 2253 | 0.858 | 0.5513 | Yes |
| 18 | IGFBP1 | 2977 | 0.618 | 0.5297 | No |
| 19 | TARS1 | 3068 | 0.595 | 0.5428 | No |
| 20 | ALDH18A1 | 3536 | 0.497 | 0.5318 | No |
| 21 | KDELR3 | 3625 | 0.480 | 0.5415 | No |
| 22 | EDEM1 | 4154 | 0.396 | 0.5241 | No |
| 23 | POP4 | 4300 | 0.378 | 0.5275 | No |
| 24 | CNOT2 | 4460 | 0.360 | 0.5295 | No |
| 25 | PARN | 4527 | 0.352 | 0.5366 | No |
| 26 | NABP1 | 4801 | 0.325 | 0.5312 | No |
| 27 | VEGFA | 4992 | 0.306 | 0.5299 | No |
| 28 | EXOSC9 | 5488 | 0.264 | 0.5103 | No |
| 29 | NHP2 | 5570 | 0.259 | 0.5136 | No |
| 30 | DCTN1 | 5859 | 0.238 | 0.5048 | No |
| 31 | PSAT1 | 5994 | 0.232 | 0.5043 | No |
| 32 | GEMIN4 | 6323 | 0.214 | 0.4925 | No |
| 33 | WIPI1 | 6416 | 0.210 | 0.4938 | No |
| 34 | TATDN2 | 6449 | 0.208 | 0.4983 | No |
| 35 | SEC31A | 6563 | 0.202 | 0.4982 | No |
| 36 | EIF2AK3 | 6739 | 0.194 | 0.4943 | No |
| 37 | EXOSC5 | 6800 | 0.191 | 0.4967 | No |
| 38 | NOP56 | 7232 | 0.173 | 0.4779 | No |
| 39 | PAIP1 | 7655 | 0.158 | 0.4592 | No |
| 40 | ERN1 | 7805 | 0.154 | 0.4555 | No |
| 41 | ASNS | 7942 | 0.149 | 0.4525 | No |
| 42 | SRPRB | 8107 | 0.144 | 0.4477 | No |
| 43 | EXOSC4 | 8127 | 0.143 | 0.4510 | No |
| 44 | MTREX | 8134 | 0.143 | 0.4550 | No |
| 45 | CHAC1 | 8138 | 0.143 | 0.4592 | No |
| 46 | SHC1 | 8280 | 0.138 | 0.4555 | No |
| 47 | EDC4 | 8414 | 0.134 | 0.4521 | No |
| 48 | NOLC1 | 8470 | 0.133 | 0.4531 | No |
| 49 | GOSR2 | 8529 | 0.131 | 0.4538 | No |
| 50 | DNAJC3 | 9004 | 0.119 | 0.4310 | No |
| 51 | FKBP14 | 9028 | 0.119 | 0.4333 | No |
| 52 | ATF4 | 9235 | 0.113 | 0.4252 | No |
| 53 | XPOT | 9261 | 0.112 | 0.4273 | No |
| 54 | SLC7A5 | 9265 | 0.112 | 0.4305 | No |
| 55 | SPCS1 | 9313 | 0.111 | 0.4313 | No |
| 56 | RRP9 | 9449 | 0.108 | 0.4270 | No |
| 57 | IARS1 | 9633 | 0.104 | 0.4200 | No |
| 58 | HYOU1 | 9805 | 0.101 | 0.4135 | No |
| 59 | CALR | 9889 | 0.099 | 0.4118 | No |
| 60 | ERO1A | 10085 | 0.095 | 0.4038 | No |
| 61 | ZBTB17 | 10367 | 0.091 | 0.3909 | No |
| 62 | XBP1 | 10646 | 0.086 | 0.3780 | No |
| 63 | NOP14 | 10798 | 0.083 | 0.3721 | No |
| 64 | SPCS3 | 11144 | 0.077 | 0.3551 | No |
| 65 | NFYA | 11152 | 0.077 | 0.3571 | No |
| 66 | SLC30A5 | 11162 | 0.077 | 0.3589 | No |
| 67 | PDIA6 | 11195 | 0.076 | 0.3594 | No |
| 68 | ARFGAP1 | 11514 | 0.071 | 0.3438 | No |
| 69 | LSM4 | 12159 | 0.062 | 0.3097 | No |
| 70 | DDX10 | 12168 | 0.062 | 0.3112 | No |
| 71 | ATF3 | 12813 | 0.053 | 0.2768 | No |
| 72 | BAG3 | 12923 | 0.051 | 0.2723 | No |
| 73 | DCP1A | 13002 | 0.050 | 0.2695 | No |
| 74 | DNAJB9 | 13003 | 0.050 | 0.2710 | No |
| 75 | LSM1 | 13213 | 0.048 | 0.2608 | No |
| 76 | SEC11A | 13548 | 0.043 | 0.2434 | No |
| 77 | HSPA5 | 13713 | 0.041 | 0.2355 | No |
| 78 | HERPUD1 | 13731 | 0.041 | 0.2358 | No |
| 79 | EIF2S1 | 14277 | 0.029 | 0.2062 | No |
| 80 | RPS14 | 14490 | 0.017 | 0.1949 | No |
| 81 | KHSRP | 14829 | -0.000 | 0.1760 | No |
| 82 | FUS | 14845 | -0.000 | 0.1751 | No |
| 83 | CCL2 | 15158 | -0.000 | 0.1577 | No |
| 84 | WFS1 | 15183 | -0.000 | 0.1564 | No |
| 85 | HSPA9 | 15260 | -0.000 | 0.1521 | No |
| 86 | CNOT6 | 15267 | -0.000 | 0.1518 | No |
| 87 | STC2 | 15284 | -0.000 | 0.1509 | No |
| 88 | SLC1A4 | 15334 | -0.000 | 0.1482 | No |
| 89 | SERP1 | 15451 | -0.000 | 0.1417 | No |
| 90 | NFYB | 15454 | -0.000 | 0.1416 | No |
| 91 | SSR1 | 15548 | -0.000 | 0.1364 | No |
| 92 | DKC1 | 15691 | -0.000 | 0.1284 | No |
| 93 | TUBB2A | 15884 | -0.000 | 0.1177 | No |
| 94 | CEBPG | 16231 | -0.000 | 0.0984 | No |
| 95 | CXXC1 | 16253 | -0.000 | 0.0972 | No |
| 96 | EIF4A2 | 16288 | -0.000 | 0.0953 | No |
| 97 | ATP6V0D1 | 16360 | -0.000 | 0.0913 | No |
| 98 | EIF4A1 | 16401 | -0.000 | 0.0891 | No |
| 99 | EEF2 | 16659 | -0.000 | 0.0748 | No |
| 100 | CEBPB | 16869 | -0.000 | 0.0631 | No |
| 101 | DCP2 | 16887 | -0.000 | 0.0621 | No |
| 102 | CKS1B | 16905 | -0.000 | 0.0612 | No |
| 103 | NPM1 | 17120 | -0.000 | 0.0492 | No |
| 104 | EIF4EBP1 | 17362 | -0.000 | 0.0358 | No |
| 105 | H2AX | 17383 | -0.000 | 0.0346 | No |