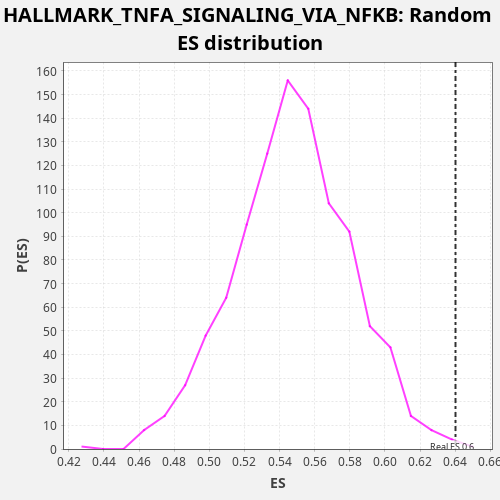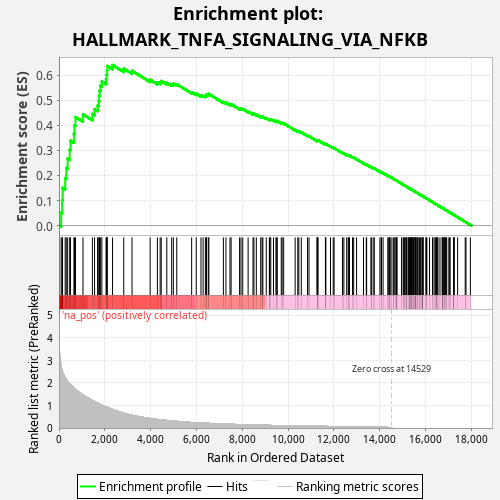
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_TNFA_SIGNALING_VIA_NFKB |
| Enrichment Score (ES) | 0.64019465 |
| Normalized Enrichment Score (NES) | 1.169455 |
| Nominal p-value | 0.004 |
| FDR q-value | 0.1378341 |
| FWER p-Value | 0.578 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | STAT5A | 93 | 2.789 | 0.0509 | Yes |
| 2 | PFKFB3 | 134 | 2.603 | 0.1010 | Yes |
| 3 | TANK | 155 | 2.510 | 0.1504 | Yes |
| 4 | NFKBIA | 270 | 2.233 | 0.1889 | Yes |
| 5 | TNFAIP8 | 316 | 2.154 | 0.2297 | Yes |
| 6 | MAFF | 378 | 2.060 | 0.2678 | Yes |
| 7 | F3 | 467 | 1.956 | 0.3022 | Yes |
| 8 | REL | 504 | 1.917 | 0.3387 | Yes |
| 9 | MSC | 651 | 1.781 | 0.3664 | Yes |
| 10 | TNFAIP2 | 677 | 1.754 | 0.4003 | Yes |
| 11 | IL6ST | 721 | 1.721 | 0.4325 | Yes |
| 12 | ZBTB10 | 1047 | 1.465 | 0.4437 | Yes |
| 13 | PLEK | 1458 | 1.222 | 0.4453 | Yes |
| 14 | CCL4 | 1553 | 1.169 | 0.4635 | Yes |
| 15 | PER1 | 1683 | 1.105 | 0.4785 | Yes |
| 16 | FJX1 | 1739 | 1.078 | 0.4971 | Yes |
| 17 | PDLIM5 | 1754 | 1.073 | 0.5179 | Yes |
| 18 | CXCL2 | 1775 | 1.064 | 0.5382 | Yes |
| 19 | TNIP2 | 1805 | 1.051 | 0.5578 | Yes |
| 20 | PTX3 | 1870 | 1.019 | 0.5747 | Yes |
| 21 | RCAN1 | 2055 | 0.944 | 0.5833 | Yes |
| 22 | TNIP1 | 2074 | 0.934 | 0.6011 | Yes |
| 23 | LDLR | 2097 | 0.924 | 0.6185 | Yes |
| 24 | IL1A | 2106 | 0.921 | 0.6365 | Yes |
| 25 | KYNU | 2338 | 0.826 | 0.6402 | Yes |
| 26 | BTG3 | 2826 | 0.655 | 0.6260 | No |
| 27 | GEM | 3188 | 0.569 | 0.6172 | No |
| 28 | TRIP10 | 3979 | 0.423 | 0.5814 | No |
| 29 | SERPINB8 | 4295 | 0.379 | 0.5713 | No |
| 30 | NFKBIE | 4416 | 0.365 | 0.5719 | No |
| 31 | NFKB2 | 4462 | 0.359 | 0.5766 | No |
| 32 | SQSTM1 | 4706 | 0.334 | 0.5697 | No |
| 33 | ABCA1 | 4919 | 0.313 | 0.5641 | No |
| 34 | VEGFA | 4992 | 0.306 | 0.5662 | No |
| 35 | PPP1R15A | 5141 | 0.293 | 0.5638 | No |
| 36 | GADD45B | 5791 | 0.243 | 0.5323 | No |
| 37 | SLC2A3 | 5992 | 0.232 | 0.5257 | No |
| 38 | CCL5 | 6199 | 0.221 | 0.5186 | No |
| 39 | IFIH1 | 6292 | 0.216 | 0.5178 | No |
| 40 | EHD1 | 6391 | 0.211 | 0.5165 | No |
| 41 | RNF19B | 6434 | 0.209 | 0.5183 | No |
| 42 | NR4A1 | 6436 | 0.209 | 0.5225 | No |
| 43 | F2RL1 | 6516 | 0.205 | 0.5222 | No |
| 44 | LAMB3 | 6542 | 0.203 | 0.5249 | No |
| 45 | TAP1 | 7176 | 0.175 | 0.4928 | No |
| 46 | PHLDA1 | 7288 | 0.171 | 0.4901 | No |
| 47 | SERPINB2 | 7465 | 0.165 | 0.4835 | No |
| 48 | CXCL6 | 7513 | 0.163 | 0.4842 | No |
| 49 | CLCF1 | 7883 | 0.152 | 0.4665 | No |
| 50 | SLC2A6 | 7945 | 0.149 | 0.4661 | No |
| 51 | PLAU | 8021 | 0.147 | 0.4648 | No |
| 52 | ICAM1 | 8256 | 0.139 | 0.4545 | No |
| 53 | TNFRSF9 | 8469 | 0.133 | 0.4452 | No |
| 54 | IL12B | 8510 | 0.132 | 0.4456 | No |
| 55 | SDC4 | 8614 | 0.129 | 0.4424 | No |
| 56 | FOSL1 | 8806 | 0.124 | 0.4342 | No |
| 57 | PANX1 | 8865 | 0.123 | 0.4334 | No |
| 58 | IL7R | 8900 | 0.122 | 0.4340 | No |
| 59 | KDM6B | 9048 | 0.118 | 0.4281 | No |
| 60 | CDKN1A | 9183 | 0.114 | 0.4229 | No |
| 61 | FUT4 | 9198 | 0.114 | 0.4244 | No |
| 62 | BIRC2 | 9245 | 0.113 | 0.4241 | No |
| 63 | OLR1 | 9363 | 0.110 | 0.4197 | No |
| 64 | DENND5A | 9478 | 0.108 | 0.4155 | No |
| 65 | DNAJB4 | 9483 | 0.108 | 0.4174 | No |
| 66 | IL15RA | 9529 | 0.107 | 0.4170 | No |
| 67 | IFNGR2 | 9703 | 0.103 | 0.4094 | No |
| 68 | TNC | 9741 | 0.102 | 0.4094 | No |
| 69 | SPHK1 | 9808 | 0.100 | 0.4077 | No |
| 70 | NFE2L2 | 10309 | 0.091 | 0.3815 | No |
| 71 | DUSP5 | 10424 | 0.090 | 0.3769 | No |
| 72 | AREG | 10470 | 0.089 | 0.3761 | No |
| 73 | BTG2 | 10578 | 0.087 | 0.3719 | No |
| 74 | NFAT5 | 10850 | 0.082 | 0.3583 | No |
| 75 | EDN1 | 10914 | 0.081 | 0.3564 | No |
| 76 | SLC16A6 | 11270 | 0.075 | 0.3380 | No |
| 77 | TIPARP | 11271 | 0.075 | 0.3395 | No |
| 78 | TRAF1 | 11280 | 0.075 | 0.3405 | No |
| 79 | CD83 | 11310 | 0.074 | 0.3404 | No |
| 80 | CD69 | 11632 | 0.069 | 0.3238 | No |
| 81 | SGK1 | 11633 | 0.069 | 0.3252 | No |
| 82 | MCL1 | 11651 | 0.069 | 0.3256 | No |
| 83 | PHLDA2 | 11853 | 0.066 | 0.3156 | No |
| 84 | IRS2 | 11971 | 0.064 | 0.3104 | No |
| 85 | PLAUR | 12009 | 0.064 | 0.3096 | No |
| 86 | IL23A | 12376 | 0.059 | 0.2902 | No |
| 87 | YRDC | 12436 | 0.058 | 0.2881 | No |
| 88 | RIPK2 | 12559 | 0.056 | 0.2823 | No |
| 89 | TRIB1 | 12625 | 0.055 | 0.2798 | No |
| 90 | PNRC1 | 12643 | 0.055 | 0.2800 | No |
| 91 | DUSP2 | 12681 | 0.055 | 0.2790 | No |
| 92 | ATF3 | 12813 | 0.053 | 0.2727 | No |
| 93 | GFPT2 | 12864 | 0.052 | 0.2709 | No |
| 94 | BIRC3 | 12991 | 0.051 | 0.2649 | No |
| 95 | CXCL1 | 13293 | 0.047 | 0.2489 | No |
| 96 | BCL2A1 | 13414 | 0.045 | 0.2431 | No |
| 97 | KLF2 | 13430 | 0.045 | 0.2432 | No |
| 98 | DRAM1 | 13623 | 0.042 | 0.2332 | No |
| 99 | SOD2 | 13630 | 0.042 | 0.2337 | No |
| 100 | B4GALT1 | 13714 | 0.041 | 0.2299 | No |
| 101 | KLF10 | 13766 | 0.040 | 0.2278 | No |
| 102 | PTGER4 | 14010 | 0.036 | 0.2149 | No |
| 103 | IER2 | 14066 | 0.035 | 0.2125 | No |
| 104 | PLK2 | 14080 | 0.035 | 0.2125 | No |
| 105 | DUSP1 | 14149 | 0.033 | 0.2093 | No |
| 106 | INHBA | 14354 | 0.026 | 0.1984 | No |
| 107 | BTG1 | 14396 | 0.022 | 0.1966 | No |
| 108 | KLF6 | 14423 | 0.021 | 0.1955 | No |
| 109 | BMP2 | 14448 | 0.020 | 0.1946 | No |
| 110 | NR4A2 | 14450 | 0.020 | 0.1949 | No |
| 111 | CFLAR | 14534 | -0.000 | 0.1903 | No |
| 112 | CD44 | 14614 | -0.000 | 0.1858 | No |
| 113 | MAP2K3 | 14622 | -0.000 | 0.1854 | No |
| 114 | MXD1 | 14670 | -0.000 | 0.1828 | No |
| 115 | BCL3 | 14715 | -0.000 | 0.1803 | No |
| 116 | ATP2B1 | 14728 | -0.000 | 0.1796 | No |
| 117 | PTGS2 | 14745 | -0.000 | 0.1787 | No |
| 118 | FOSL2 | 14755 | -0.000 | 0.1782 | No |
| 119 | JAG1 | 14955 | -0.000 | 0.1671 | No |
| 120 | TSC22D1 | 15023 | -0.000 | 0.1633 | No |
| 121 | RELB | 15052 | -0.000 | 0.1617 | No |
| 122 | NAMPT | 15086 | -0.000 | 0.1599 | No |
| 123 | MAP3K8 | 15139 | -0.000 | 0.1570 | No |
| 124 | CCL2 | 15158 | -0.000 | 0.1560 | No |
| 125 | NFKB1 | 15179 | -0.000 | 0.1548 | No |
| 126 | CCND1 | 15191 | -0.000 | 0.1542 | No |
| 127 | HBEGF | 15262 | -0.000 | 0.1503 | No |
| 128 | BCL6 | 15287 | -0.000 | 0.1489 | No |
| 129 | HES1 | 15293 | -0.000 | 0.1487 | No |
| 130 | CCL20 | 15302 | -0.000 | 0.1482 | No |
| 131 | ID2 | 15330 | -0.000 | 0.1467 | No |
| 132 | GADD45A | 15351 | -0.000 | 0.1456 | No |
| 133 | TNFAIP3 | 15390 | -0.000 | 0.1434 | No |
| 134 | KLF9 | 15401 | -0.000 | 0.1429 | No |
| 135 | NR4A3 | 15407 | -0.000 | 0.1426 | No |
| 136 | EGR1 | 15450 | -0.000 | 0.1402 | No |
| 137 | DUSP4 | 15455 | -0.000 | 0.1400 | No |
| 138 | CD80 | 15469 | -0.000 | 0.1393 | No |
| 139 | EGR2 | 15499 | -0.000 | 0.1377 | No |
| 140 | PMEPA1 | 15538 | -0.000 | 0.1355 | No |
| 141 | IRF1 | 15558 | -0.000 | 0.1345 | No |
| 142 | IL1B | 15568 | -0.000 | 0.1339 | No |
| 143 | TNFSF9 | 15571 | -0.000 | 0.1338 | No |
| 144 | FOSB | 15574 | -0.000 | 0.1337 | No |
| 145 | ZFP36 | 15627 | -0.000 | 0.1308 | No |
| 146 | LIF | 15635 | -0.000 | 0.1304 | No |
| 147 | SAT1 | 15668 | -0.000 | 0.1286 | No |
| 148 | NINJ1 | 15718 | -0.000 | 0.1259 | No |
| 149 | GCH1 | 15732 | -0.000 | 0.1251 | No |
| 150 | BHLHE40 | 15778 | -0.000 | 0.1226 | No |
| 151 | IL6 | 15843 | -0.000 | 0.1190 | No |
| 152 | KLF4 | 15860 | -0.000 | 0.1181 | No |
| 153 | MYC | 15873 | -0.000 | 0.1174 | No |
| 154 | TUBB2A | 15884 | -0.000 | 0.1169 | No |
| 155 | IER3 | 15886 | -0.000 | 0.1168 | No |
| 156 | TLR2 | 15888 | -0.000 | 0.1168 | No |
| 157 | CCN1 | 16014 | -0.000 | 0.1098 | No |
| 158 | RHOB | 16045 | -0.000 | 0.1081 | No |
| 159 | ACKR3 | 16059 | -0.000 | 0.1073 | No |
| 160 | IL18 | 16176 | -0.000 | 0.1008 | No |
| 161 | ETS2 | 16305 | -0.000 | 0.0936 | No |
| 162 | B4GALT5 | 16327 | -0.000 | 0.0925 | No |
| 163 | PLPP3 | 16410 | -0.000 | 0.0879 | No |
| 164 | IER5 | 16427 | -0.000 | 0.0870 | No |
| 165 | CCNL1 | 16471 | -0.000 | 0.0845 | No |
| 166 | CXCL3 | 16475 | -0.000 | 0.0844 | No |
| 167 | ZC3H12A | 16482 | -0.000 | 0.0840 | No |
| 168 | CSF2 | 16515 | -0.000 | 0.0822 | No |
| 169 | NFIL3 | 16546 | -0.000 | 0.0806 | No |
| 170 | SMAD3 | 16632 | -0.000 | 0.0758 | No |
| 171 | EFNA1 | 16735 | -0.000 | 0.0701 | No |
| 172 | CXCL10 | 16736 | -0.000 | 0.0701 | No |
| 173 | CXCL11 | 16737 | -0.000 | 0.0701 | No |
| 174 | GPR183 | 16748 | -0.000 | 0.0695 | No |
| 175 | FOS | 16780 | -0.000 | 0.0678 | No |
| 176 | JUNB | 16825 | -0.000 | 0.0653 | No |
| 177 | SPSB1 | 16847 | -0.000 | 0.0641 | No |
| 178 | CEBPB | 16869 | -0.000 | 0.0629 | No |
| 179 | RELA | 16902 | -0.000 | 0.0611 | No |
| 180 | EIF1 | 16928 | -0.000 | 0.0597 | No |
| 181 | TGIF1 | 17025 | -0.000 | 0.0543 | No |
| 182 | JUN | 17035 | -0.000 | 0.0538 | No |
| 183 | EGR3 | 17076 | -0.000 | 0.0516 | No |
| 184 | CSF1 | 17230 | -0.000 | 0.0430 | No |
| 185 | SOCS3 | 17241 | -0.000 | 0.0424 | No |
| 186 | PDE4B | 17242 | -0.000 | 0.0424 | No |
| 187 | SNN | 17243 | -0.000 | 0.0424 | No |
| 188 | LITAF | 17402 | -0.000 | 0.0336 | No |
| 189 | CEBPD | 17733 | -0.000 | 0.0150 | No |
| 190 | TNF | 17767 | -0.000 | 0.0132 | No |
| 191 | MARCKS | 17959 | -0.000 | 0.0025 | No |