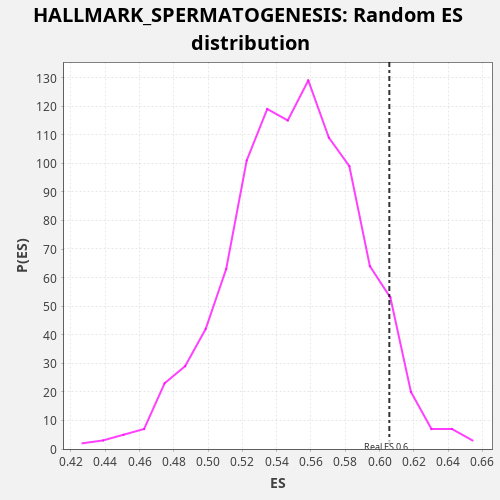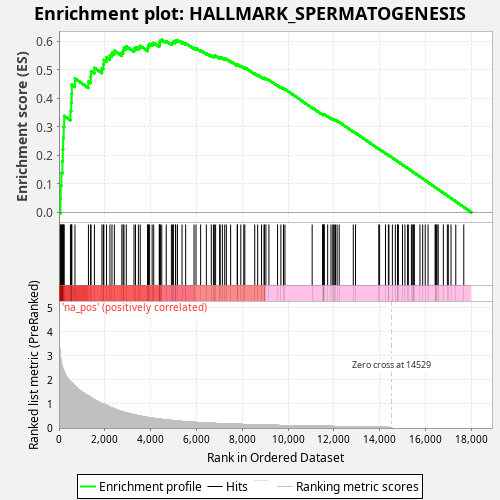
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.60573155 |
| Normalized Enrichment Score (NES) | 1.1006372 |
| Nominal p-value | 0.061 |
| FDR q-value | 0.35750073 |
| FWER p-Value | 0.993 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | HSPA4L | 60 | 2.980 | 0.0470 | Yes |
| 2 | DDX25 | 74 | 2.893 | 0.0952 | Yes |
| 3 | NCAPH | 107 | 2.692 | 0.1389 | Yes |
| 4 | BRAF | 149 | 2.541 | 0.1795 | Yes |
| 5 | GAD1 | 165 | 2.483 | 0.2207 | Yes |
| 6 | CSNK2A2 | 181 | 2.440 | 0.2611 | Yes |
| 7 | AGFG1 | 204 | 2.381 | 0.3001 | Yes |
| 8 | TTK | 226 | 2.324 | 0.3382 | Yes |
| 9 | NAA11 | 500 | 1.920 | 0.3553 | Yes |
| 10 | MTOR | 534 | 1.894 | 0.3855 | Yes |
| 11 | H1-6 | 550 | 1.879 | 0.4164 | Yes |
| 12 | DCC | 559 | 1.871 | 0.4476 | Yes |
| 13 | RAD17 | 697 | 1.738 | 0.4693 | Yes |
| 14 | ADAD1 | 1285 | 1.328 | 0.4589 | Yes |
| 15 | CCT6B | 1381 | 1.268 | 0.4750 | Yes |
| 16 | SLC12A2 | 1397 | 1.260 | 0.4955 | Yes |
| 17 | CAMK4 | 1546 | 1.172 | 0.5070 | Yes |
| 18 | DBF4 | 1876 | 1.017 | 0.5058 | Yes |
| 19 | PRKAR2A | 1936 | 0.989 | 0.5192 | Yes |
| 20 | CFTR | 1959 | 0.979 | 0.5345 | Yes |
| 21 | LPIN1 | 2075 | 0.933 | 0.5438 | Yes |
| 22 | ACRV1 | 2223 | 0.871 | 0.5503 | Yes |
| 23 | ZC3H14 | 2312 | 0.833 | 0.5595 | Yes |
| 24 | MEP1B | 2416 | 0.796 | 0.5671 | Yes |
| 25 | SHE | 2744 | 0.677 | 0.5603 | Yes |
| 26 | JAM3 | 2800 | 0.661 | 0.5684 | Yes |
| 27 | PEBP1 | 2836 | 0.653 | 0.5774 | Yes |
| 28 | ZC2HC1C | 2937 | 0.627 | 0.5825 | Yes |
| 29 | TALDO1 | 3273 | 0.548 | 0.5730 | Yes |
| 30 | LDHC | 3339 | 0.535 | 0.5784 | Yes |
| 31 | TCP11 | 3466 | 0.511 | 0.5800 | Yes |
| 32 | SCG5 | 3546 | 0.495 | 0.5839 | Yes |
| 33 | TNP2 | 3867 | 0.439 | 0.5734 | Yes |
| 34 | TEKT2 | 3884 | 0.436 | 0.5799 | Yes |
| 35 | MAP7 | 3907 | 0.432 | 0.5860 | Yes |
| 36 | AURKA | 3956 | 0.426 | 0.5905 | Yes |
| 37 | GAPDHS | 4066 | 0.411 | 0.5913 | Yes |
| 38 | ACRBP | 4135 | 0.400 | 0.5943 | Yes |
| 39 | HBZ | 4378 | 0.369 | 0.5869 | Yes |
| 40 | IFT88 | 4387 | 0.368 | 0.5927 | Yes |
| 41 | ODF1 | 4398 | 0.367 | 0.5984 | Yes |
| 42 | DPEP3 | 4426 | 0.363 | 0.6030 | Yes |
| 43 | PRM2 | 4486 | 0.357 | 0.6057 | Yes |
| 44 | ADAM2 | 4681 | 0.337 | 0.6006 | No |
| 45 | ACE | 4906 | 0.314 | 0.5933 | No |
| 46 | NEFH | 4958 | 0.310 | 0.5957 | No |
| 47 | PGS1 | 4993 | 0.306 | 0.5990 | No |
| 48 | CHFR | 5084 | 0.299 | 0.5990 | No |
| 49 | TULP2 | 5095 | 0.298 | 0.6035 | No |
| 50 | CST8 | 5171 | 0.291 | 0.6042 | No |
| 51 | MLF1 | 5371 | 0.273 | 0.5977 | No |
| 52 | POMC | 5529 | 0.261 | 0.5933 | No |
| 53 | NPHP1 | 5895 | 0.237 | 0.5769 | No |
| 54 | RFC4 | 5981 | 0.232 | 0.5760 | No |
| 55 | SPATA6 | 6183 | 0.222 | 0.5685 | No |
| 56 | ALOX15 | 6432 | 0.209 | 0.5582 | No |
| 57 | SLC2A5 | 6648 | 0.198 | 0.5495 | No |
| 58 | CCNB2 | 6741 | 0.193 | 0.5476 | No |
| 59 | HOXB1 | 6781 | 0.192 | 0.5487 | No |
| 60 | SYCP1 | 6837 | 0.189 | 0.5488 | No |
| 61 | BUB1 | 7013 | 0.182 | 0.5421 | No |
| 62 | SCG3 | 7036 | 0.181 | 0.5439 | No |
| 63 | NEK2 | 7132 | 0.177 | 0.5416 | No |
| 64 | OAZ3 | 7235 | 0.173 | 0.5388 | No |
| 65 | GRM8 | 7305 | 0.171 | 0.5378 | No |
| 66 | GSTM3 | 7493 | 0.164 | 0.5302 | No |
| 67 | TUBA3C | 7784 | 0.155 | 0.5165 | No |
| 68 | ART3 | 7786 | 0.155 | 0.5191 | No |
| 69 | KIF2C | 7937 | 0.149 | 0.5132 | No |
| 70 | HSPA1L | 8066 | 0.145 | 0.5085 | No |
| 71 | PHF7 | 8112 | 0.144 | 0.5084 | No |
| 72 | DDX4 | 8545 | 0.131 | 0.4865 | No |
| 73 | CDKN3 | 8673 | 0.127 | 0.4815 | No |
| 74 | TNNI3 | 8842 | 0.123 | 0.4742 | No |
| 75 | CLGN | 8955 | 0.120 | 0.4700 | No |
| 76 | NOS1 | 8963 | 0.120 | 0.4716 | No |
| 77 | PAPOLB | 9033 | 0.118 | 0.4697 | No |
| 78 | CLPB | 9165 | 0.115 | 0.4643 | No |
| 79 | PGK2 | 9542 | 0.106 | 0.4451 | No |
| 80 | PARP2 | 9687 | 0.103 | 0.4388 | No |
| 81 | STAM2 | 9800 | 0.101 | 0.4342 | No |
| 82 | ACTL7B | 9863 | 0.099 | 0.4324 | No |
| 83 | GMCL1 | 11052 | 0.078 | 0.3673 | No |
| 84 | PHKG2 | 11506 | 0.072 | 0.3432 | No |
| 85 | TLE4 | 11538 | 0.071 | 0.3426 | No |
| 86 | HTR5A | 11555 | 0.071 | 0.3429 | No |
| 87 | MLLT10 | 11562 | 0.071 | 0.3438 | No |
| 88 | PCSK4 | 11579 | 0.070 | 0.3441 | No |
| 89 | MTNR1A | 11729 | 0.068 | 0.3369 | No |
| 90 | ELOVL3 | 11873 | 0.066 | 0.3300 | No |
| 91 | SEPTIN4 | 11952 | 0.064 | 0.3267 | No |
| 92 | TSSK2 | 11999 | 0.064 | 0.3252 | No |
| 93 | SIRT1 | 12011 | 0.064 | 0.3257 | No |
| 94 | TOPBP1 | 12072 | 0.063 | 0.3234 | No |
| 95 | PSMG1 | 12141 | 0.062 | 0.3206 | No |
| 96 | CRISP2 | 12232 | 0.061 | 0.3166 | No |
| 97 | ZPBP | 12847 | 0.052 | 0.2831 | No |
| 98 | DMC1 | 12945 | 0.051 | 0.2786 | No |
| 99 | CLVS1 | 13960 | 0.037 | 0.2225 | No |
| 100 | PIAS2 | 13996 | 0.036 | 0.2211 | No |
| 101 | TKTL1 | 14253 | 0.031 | 0.2073 | No |
| 102 | DNAJB8 | 14379 | 0.024 | 0.2007 | No |
| 103 | ZNRF4 | 14403 | 0.022 | 0.1998 | No |
| 104 | YBX2 | 14554 | -0.000 | 0.1914 | No |
| 105 | SNAP91 | 14683 | -0.000 | 0.1843 | No |
| 106 | VDAC3 | 14776 | -0.000 | 0.1791 | No |
| 107 | IL12RB2 | 14793 | -0.000 | 0.1782 | No |
| 108 | MAST2 | 14813 | -0.000 | 0.1771 | No |
| 109 | PCSK1N | 14997 | -0.000 | 0.1669 | No |
| 110 | EZH2 | 15104 | -0.000 | 0.1610 | No |
| 111 | GSG1 | 15218 | -0.000 | 0.1547 | No |
| 112 | PACRG | 15251 | -0.000 | 0.1529 | No |
| 113 | TNP1 | 15383 | -0.000 | 0.1455 | No |
| 114 | IDE | 15419 | -0.000 | 0.1436 | No |
| 115 | COIL | 15462 | -0.000 | 0.1412 | No |
| 116 | ARL4A | 15491 | -0.000 | 0.1397 | No |
| 117 | IL13RA2 | 15511 | -0.000 | 0.1386 | No |
| 118 | CCNA1 | 15758 | -0.000 | 0.1248 | No |
| 119 | DMRT1 | 15877 | -0.000 | 0.1182 | No |
| 120 | ADCYAP1 | 15988 | -0.000 | 0.1121 | No |
| 121 | AKAP4 | 16111 | -0.000 | 0.1052 | No |
| 122 | GFI1 | 16419 | -0.000 | 0.0881 | No |
| 123 | PDHA2 | 16439 | -0.000 | 0.0870 | No |
| 124 | RPL39L | 16486 | -0.000 | 0.0844 | No |
| 125 | NPY5R | 16501 | -0.000 | 0.0837 | No |
| 126 | STRBP | 16555 | -0.000 | 0.0807 | No |
| 127 | CDK1 | 16779 | -0.000 | 0.0682 | No |
| 128 | CNIH2 | 16955 | -0.000 | 0.0584 | No |
| 129 | IP6K1 | 16995 | -0.000 | 0.0562 | No |
| 130 | CHRM4 | 17114 | -0.000 | 0.0496 | No |
| 131 | NF2 | 17322 | -0.000 | 0.0380 | No |
| 132 | TSN | 17671 | -0.000 | 0.0186 | No |