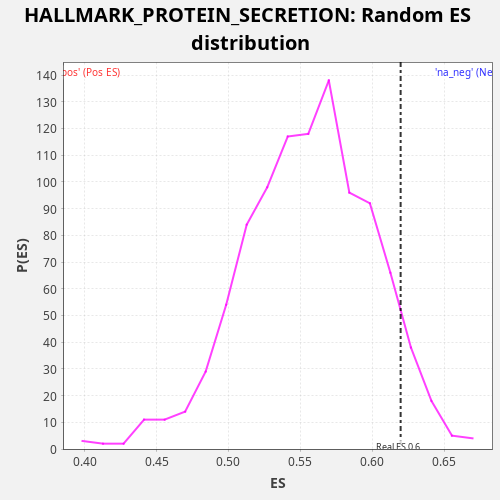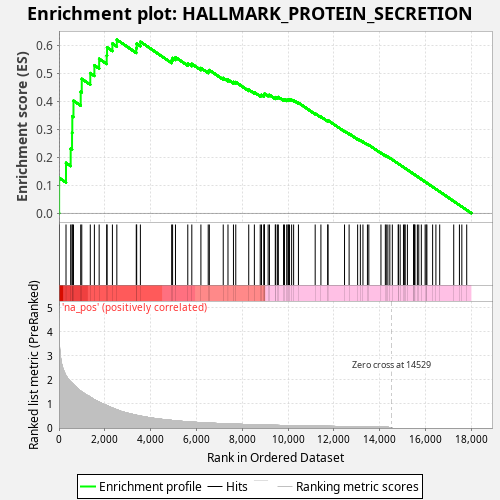
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.6197017 |
| Normalized Enrichment Score (NES) | 1.1162606 |
| Nominal p-value | 0.065 |
| FDR q-value | 0.29520196 |
| FWER p-Value | 0.945 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | TSG101 | 6 | 3.922 | 0.1265 | Yes |
| 2 | VAMP3 | 306 | 2.165 | 0.1799 | Yes |
| 3 | RPS6KA3 | 510 | 1.911 | 0.2303 | Yes |
| 4 | TMED10 | 570 | 1.861 | 0.2873 | Yes |
| 5 | TPD52 | 581 | 1.850 | 0.3465 | Yes |
| 6 | CLCN3 | 632 | 1.796 | 0.4018 | Yes |
| 7 | DST | 948 | 1.531 | 0.4338 | Yes |
| 8 | STX12 | 994 | 1.500 | 0.4798 | Yes |
| 9 | SH3GL2 | 1366 | 1.274 | 0.5003 | Yes |
| 10 | NAPA | 1545 | 1.172 | 0.5283 | Yes |
| 11 | BNIP3 | 1753 | 1.073 | 0.5514 | Yes |
| 12 | AP3B1 | 2083 | 0.930 | 0.5632 | Yes |
| 13 | DOP1A | 2098 | 0.924 | 0.5923 | Yes |
| 14 | GBF1 | 2332 | 0.828 | 0.6060 | Yes |
| 15 | ZW10 | 2525 | 0.754 | 0.6197 | Yes |
| 16 | TOM1L1 | 3373 | 0.528 | 0.5895 | No |
| 17 | BET1 | 3391 | 0.525 | 0.6055 | No |
| 18 | CTSC | 3553 | 0.494 | 0.6125 | No |
| 19 | ABCA1 | 4919 | 0.313 | 0.5464 | No |
| 20 | CLN5 | 4959 | 0.310 | 0.5543 | No |
| 21 | GALC | 5085 | 0.299 | 0.5569 | No |
| 22 | SEC24D | 5622 | 0.255 | 0.5353 | No |
| 23 | GNAS | 5798 | 0.243 | 0.5333 | No |
| 24 | ATP6V1B1 | 6195 | 0.221 | 0.5184 | No |
| 25 | MON2 | 6514 | 0.205 | 0.5072 | No |
| 26 | SEC31A | 6563 | 0.202 | 0.5111 | No |
| 27 | VPS45 | 7170 | 0.175 | 0.4829 | No |
| 28 | SCRN1 | 7377 | 0.168 | 0.4769 | No |
| 29 | SSPN | 7616 | 0.160 | 0.4688 | No |
| 30 | SCAMP3 | 7717 | 0.157 | 0.4682 | No |
| 31 | PAM | 8283 | 0.138 | 0.4412 | No |
| 32 | GOSR2 | 8529 | 0.131 | 0.4317 | No |
| 33 | KIF1B | 8785 | 0.125 | 0.4215 | No |
| 34 | COG2 | 8840 | 0.123 | 0.4225 | No |
| 35 | ATP6V1H | 8947 | 0.120 | 0.4205 | No |
| 36 | LMAN1 | 8952 | 0.120 | 0.4241 | No |
| 37 | STX16 | 8971 | 0.120 | 0.4270 | No |
| 38 | ARFGEF2 | 9130 | 0.116 | 0.4219 | No |
| 39 | AP3S1 | 9188 | 0.114 | 0.4224 | No |
| 40 | DNM1L | 9442 | 0.108 | 0.4118 | No |
| 41 | GOLGA4 | 9460 | 0.108 | 0.4143 | No |
| 42 | ARFGEF1 | 9534 | 0.107 | 0.4137 | No |
| 43 | EGFR | 9576 | 0.106 | 0.4148 | No |
| 44 | ICA1 | 9802 | 0.101 | 0.4055 | No |
| 45 | ERGIC3 | 9844 | 0.100 | 0.4065 | No |
| 46 | VPS4B | 9941 | 0.098 | 0.4043 | No |
| 47 | ARFIP1 | 9995 | 0.097 | 0.4044 | No |
| 48 | NAPG | 10017 | 0.097 | 0.4064 | No |
| 49 | TMX1 | 10064 | 0.096 | 0.4069 | No |
| 50 | M6PR | 10156 | 0.094 | 0.4049 | No |
| 51 | USO1 | 10243 | 0.093 | 0.4031 | No |
| 52 | COPE | 10453 | 0.089 | 0.3943 | No |
| 53 | AP2S1 | 11184 | 0.076 | 0.3560 | No |
| 54 | STX7 | 11434 | 0.073 | 0.3445 | No |
| 55 | SNX2 | 11730 | 0.068 | 0.3302 | No |
| 56 | SOD1 | 11745 | 0.068 | 0.3316 | No |
| 57 | AP2M1 | 12465 | 0.058 | 0.2933 | No |
| 58 | TSPAN8 | 12665 | 0.055 | 0.2840 | No |
| 59 | VAMP4 | 13037 | 0.050 | 0.2649 | No |
| 60 | SNAP23 | 13156 | 0.048 | 0.2599 | No |
| 61 | SEC22B | 13269 | 0.047 | 0.2551 | No |
| 62 | STAM | 13465 | 0.044 | 0.2457 | No |
| 63 | RAB22A | 13526 | 0.043 | 0.2437 | No |
| 64 | ADAM10 | 14055 | 0.035 | 0.2154 | No |
| 65 | AP2B1 | 14247 | 0.031 | 0.2057 | No |
| 66 | GLA | 14287 | 0.029 | 0.2045 | No |
| 67 | YIPF6 | 14357 | 0.026 | 0.2015 | No |
| 68 | ATP7A | 14443 | 0.020 | 0.1974 | No |
| 69 | LAMP2 | 14552 | -0.000 | 0.1913 | No |
| 70 | SCAMP1 | 14812 | -0.000 | 0.1769 | No |
| 71 | TMED2 | 14815 | -0.000 | 0.1768 | No |
| 72 | MAPK1 | 14894 | -0.000 | 0.1724 | No |
| 73 | RAB2A | 15038 | -0.000 | 0.1644 | No |
| 74 | CAV2 | 15092 | -0.000 | 0.1615 | No |
| 75 | YKT6 | 15111 | -0.000 | 0.1605 | No |
| 76 | KRT18 | 15210 | -0.000 | 0.1550 | No |
| 77 | OCRL | 15478 | -0.000 | 0.1401 | No |
| 78 | CLTA | 15495 | -0.000 | 0.1392 | No |
| 79 | RAB9A | 15516 | -0.000 | 0.1381 | No |
| 80 | VAMP7 | 15541 | -0.000 | 0.1367 | No |
| 81 | COPB1 | 15645 | -0.000 | 0.1310 | No |
| 82 | PPT1 | 15706 | -0.000 | 0.1276 | No |
| 83 | CD63 | 15818 | -0.000 | 0.1214 | No |
| 84 | CLTC | 15984 | -0.000 | 0.1122 | No |
| 85 | ANP32E | 16030 | -0.000 | 0.1097 | No |
| 86 | RAB5A | 16061 | -0.000 | 0.1080 | No |
| 87 | RER1 | 16311 | -0.000 | 0.0941 | No |
| 88 | ATP1A1 | 16454 | -0.000 | 0.0862 | No |
| 89 | AP1G1 | 16620 | -0.000 | 0.0770 | No |
| 90 | COPB2 | 17232 | -0.000 | 0.0429 | No |
| 91 | IGF2R | 17481 | -0.000 | 0.0290 | No |
| 92 | SGMS1 | 17586 | -0.000 | 0.0232 | No |
| 93 | ARFGAP3 | 17799 | -0.000 | 0.0114 | No |