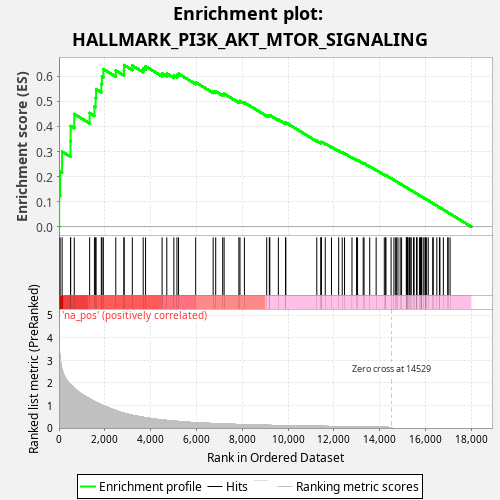
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
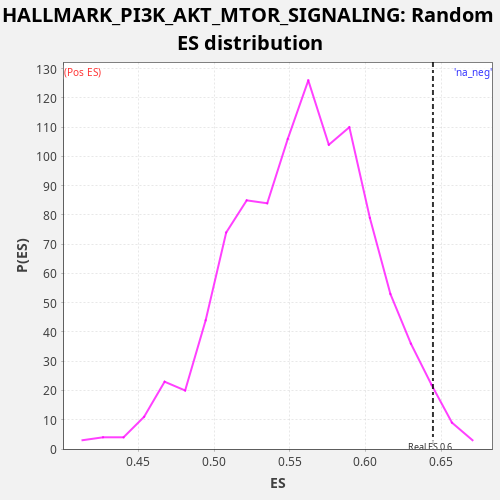
| Enrichment Score (ES) | 0.6445703 |
| Normalized Enrichment Score (NES) | 1.1566749 |
| Nominal p-value | 0.02 |
| FDR q-value | 0.15785825 |
| FWER p-Value | 0.693 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | PDK1 | 8 | 3.901 | 0.1246 | Yes |
| 2 | MKNK1 | 49 | 3.082 | 0.2212 | Yes |
| 3 | STAT2 | 136 | 2.601 | 0.2997 | Yes |
| 4 | GRK2 | 502 | 1.918 | 0.3408 | Yes |
| 5 | RPS6KA3 | 510 | 1.911 | 0.4017 | Yes |
| 6 | PPP2R1B | 666 | 1.764 | 0.4496 | Yes |
| 7 | PLA2G12A | 1338 | 1.297 | 0.4537 | Yes |
| 8 | CAMK4 | 1546 | 1.172 | 0.4797 | Yes |
| 9 | HSP90B1 | 1598 | 1.145 | 0.5136 | Yes |
| 10 | VAV3 | 1618 | 1.130 | 0.5487 | Yes |
| 11 | PRKAA2 | 1847 | 1.031 | 0.5690 | Yes |
| 12 | MYD88 | 1877 | 1.016 | 0.6000 | Yes |
| 13 | PRKAR2A | 1936 | 0.989 | 0.6285 | Yes |
| 14 | ACACA | 2479 | 0.772 | 0.6229 | Yes |
| 15 | UBE2D3 | 2835 | 0.653 | 0.6241 | Yes |
| 16 | MKNK2 | 2842 | 0.651 | 0.6446 | Yes |
| 17 | ADCY2 | 3200 | 0.565 | 0.6427 | No |
| 18 | PIKFYVE | 3675 | 0.470 | 0.6313 | No |
| 19 | TRIB3 | 3779 | 0.453 | 0.6401 | No |
| 20 | GNA14 | 4501 | 0.355 | 0.6112 | No |
| 21 | SQSTM1 | 4706 | 0.334 | 0.6105 | No |
| 22 | PAK4 | 5015 | 0.304 | 0.6031 | No |
| 23 | HRAS | 5149 | 0.293 | 0.6050 | No |
| 24 | NOD1 | 5219 | 0.286 | 0.6103 | No |
| 25 | PIK3R3 | 5967 | 0.233 | 0.5761 | No |
| 26 | ECSIT | 6729 | 0.194 | 0.5398 | No |
| 27 | CAB39L | 6841 | 0.189 | 0.5396 | No |
| 28 | LCK | 7146 | 0.176 | 0.5283 | No |
| 29 | RIT1 | 7208 | 0.174 | 0.5305 | No |
| 30 | IRAK4 | 7852 | 0.152 | 0.4995 | No |
| 31 | PLCG1 | 7900 | 0.151 | 0.5017 | No |
| 32 | AKT1S1 | 8094 | 0.144 | 0.4955 | No |
| 33 | RAC1 | 9068 | 0.117 | 0.4449 | No |
| 34 | CDKN1A | 9183 | 0.114 | 0.4422 | No |
| 35 | TIAM1 | 9204 | 0.114 | 0.4448 | No |
| 36 | EGFR | 9576 | 0.106 | 0.4274 | No |
| 37 | CALR | 9889 | 0.099 | 0.4132 | No |
| 38 | DAPP1 | 9903 | 0.099 | 0.4156 | No |
| 39 | PLCB1 | 11254 | 0.075 | 0.3426 | No |
| 40 | PPP1CA | 11432 | 0.073 | 0.3351 | No |
| 41 | TBK1 | 11445 | 0.072 | 0.3367 | No |
| 42 | RPS6KA1 | 11453 | 0.072 | 0.3387 | No |
| 43 | THEM4 | 11623 | 0.070 | 0.3315 | No |
| 44 | CDK2 | 11895 | 0.065 | 0.3184 | No |
| 45 | MAPK9 | 12210 | 0.061 | 0.3028 | No |
| 46 | MAPKAP1 | 12365 | 0.059 | 0.2961 | No |
| 47 | AP2M1 | 12465 | 0.058 | 0.2925 | No |
| 48 | MAPK8 | 12790 | 0.053 | 0.2761 | No |
| 49 | DDIT3 | 12990 | 0.051 | 0.2666 | No |
| 50 | ATF1 | 13034 | 0.050 | 0.2658 | No |
| 51 | RIPK1 | 13279 | 0.047 | 0.2536 | No |
| 52 | SLA | 13317 | 0.046 | 0.2531 | No |
| 53 | PTEN | 13565 | 0.042 | 0.2406 | No |
| 54 | FGF6 | 13845 | 0.039 | 0.2263 | No |
| 55 | CSNK2B | 14201 | 0.032 | 0.2075 | No |
| 56 | PFN1 | 14264 | 0.030 | 0.2050 | No |
| 57 | GRB2 | 14267 | 0.030 | 0.2059 | No |
| 58 | UBE2N | 14497 | 0.016 | 0.1936 | No |
| 59 | MAP2K3 | 14622 | -0.000 | 0.1866 | No |
| 60 | TNFRSF1A | 14698 | -0.000 | 0.1825 | No |
| 61 | FGF22 | 14726 | -0.000 | 0.1809 | No |
| 62 | GSK3B | 14797 | -0.000 | 0.1770 | No |
| 63 | MAPK1 | 14894 | -0.000 | 0.1717 | No |
| 64 | E2F1 | 14956 | -0.000 | 0.1683 | No |
| 65 | DUSP3 | 15164 | -0.000 | 0.1567 | No |
| 66 | MAP2K6 | 15167 | -0.000 | 0.1566 | No |
| 67 | MAPK10 | 15180 | -0.000 | 0.1559 | No |
| 68 | ARPC3 | 15214 | -0.000 | 0.1541 | No |
| 69 | CDKN1B | 15217 | -0.000 | 0.1540 | No |
| 70 | IL4 | 15275 | -0.000 | 0.1508 | No |
| 71 | ACTR3 | 15303 | -0.000 | 0.1493 | No |
| 72 | SLC2A1 | 15365 | -0.000 | 0.1459 | No |
| 73 | FASLG | 15369 | -0.000 | 0.1457 | No |
| 74 | CXCR4 | 15475 | -0.000 | 0.1398 | No |
| 75 | ITPR2 | 15503 | -0.000 | 0.1383 | No |
| 76 | TRAF2 | 15615 | -0.000 | 0.1321 | No |
| 77 | PIN1 | 15617 | -0.000 | 0.1321 | No |
| 78 | GNGT1 | 15624 | -0.000 | 0.1317 | No |
| 79 | RAF1 | 15736 | -0.000 | 0.1255 | No |
| 80 | NGF | 15783 | -0.000 | 0.1230 | No |
| 81 | MAP3K7 | 15814 | -0.000 | 0.1213 | No |
| 82 | CDK4 | 15820 | -0.000 | 0.1210 | No |
| 83 | CAB39 | 15833 | -0.000 | 0.1204 | No |
| 84 | ACTR2 | 15901 | -0.000 | 0.1166 | No |
| 85 | CLTC | 15984 | -0.000 | 0.1120 | No |
| 86 | ARHGDIA | 15991 | -0.000 | 0.1117 | No |
| 87 | RPTOR | 15992 | -0.000 | 0.1117 | No |
| 88 | RALB | 16049 | -0.000 | 0.1086 | No |
| 89 | IL2RG | 16121 | -0.000 | 0.1046 | No |
| 90 | NCK1 | 16316 | -0.000 | 0.0938 | No |
| 91 | FGF17 | 16337 | -0.000 | 0.0927 | No |
| 92 | PITX2 | 16495 | -0.000 | 0.0839 | No |
| 93 | PRKCB | 16609 | -0.000 | 0.0776 | No |
| 94 | YWHAB | 16630 | -0.000 | 0.0765 | No |
| 95 | CDK1 | 16779 | -0.000 | 0.0682 | No |
| 96 | SMAD2 | 16973 | -0.000 | 0.0574 | No |
| 97 | SFN | 16986 | -0.000 | 0.0567 | No |
| 98 | PTPN11 | 17073 | -0.000 | 0.0519 | No |