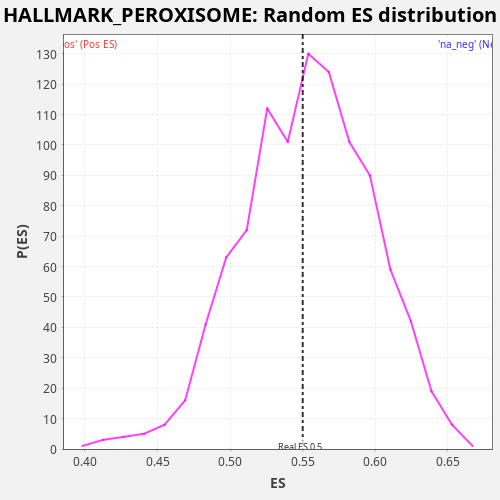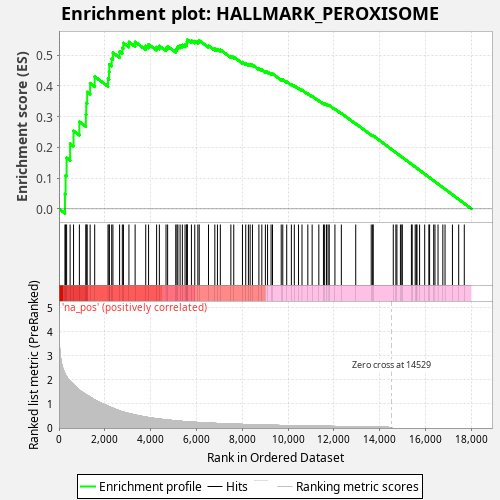
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.54999816 |
| Normalized Enrichment Score (NES) | 0.9942952 |
| Nominal p-value | 0.547 |
| FDR q-value | 0.8288477 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | SLC25A4 | 260 | 2.250 | 0.0485 | Yes |
| 2 | MSH2 | 288 | 2.197 | 0.1086 | Yes |
| 3 | CRABP1 | 332 | 2.130 | 0.1659 | Yes |
| 4 | IDI1 | 485 | 1.934 | 0.2116 | Yes |
| 5 | ELOVL5 | 635 | 1.791 | 0.2535 | Yes |
| 6 | FADS1 | 890 | 1.571 | 0.2833 | Yes |
| 7 | DIO1 | 1172 | 1.389 | 0.3065 | Yes |
| 8 | MLYCD | 1189 | 1.376 | 0.3442 | Yes |
| 9 | IDH2 | 1231 | 1.354 | 0.3799 | Yes |
| 10 | CLN6 | 1359 | 1.280 | 0.4086 | Yes |
| 11 | SLC35B2 | 1558 | 1.166 | 0.4303 | Yes |
| 12 | ABCC8 | 2139 | 0.909 | 0.4234 | Yes |
| 13 | ABCD3 | 2185 | 0.885 | 0.4457 | Yes |
| 14 | PRDX1 | 2197 | 0.883 | 0.4698 | Yes |
| 15 | ABCB4 | 2301 | 0.839 | 0.4876 | Yes |
| 16 | CRAT | 2354 | 0.819 | 0.5076 | Yes |
| 17 | CLN8 | 2645 | 0.711 | 0.5113 | Yes |
| 18 | ABCB1 | 2765 | 0.671 | 0.5235 | Yes |
| 19 | PEX2 | 2813 | 0.657 | 0.5393 | Yes |
| 20 | ALB | 3053 | 0.598 | 0.5427 | Yes |
| 21 | CAT | 3323 | 0.539 | 0.5428 | Yes |
| 22 | ESR2 | 3786 | 0.452 | 0.5296 | Yes |
| 23 | ACOT8 | 3910 | 0.432 | 0.5349 | Yes |
| 24 | HSD17B4 | 4261 | 0.382 | 0.5260 | Yes |
| 25 | ACSL5 | 4381 | 0.369 | 0.5297 | Yes |
| 26 | GSTK1 | 4672 | 0.338 | 0.5230 | Yes |
| 27 | HSD3B7 | 4742 | 0.330 | 0.5284 | Yes |
| 28 | PEX6 | 5097 | 0.298 | 0.5170 | Yes |
| 29 | HRAS | 5149 | 0.293 | 0.5223 | Yes |
| 30 | CDK7 | 5190 | 0.289 | 0.5282 | Yes |
| 31 | HMGCL | 5286 | 0.281 | 0.5307 | Yes |
| 32 | ERCC3 | 5385 | 0.272 | 0.5329 | Yes |
| 33 | HAO2 | 5503 | 0.263 | 0.5337 | Yes |
| 34 | LONP2 | 5574 | 0.258 | 0.5370 | Yes |
| 35 | TOP2A | 5588 | 0.258 | 0.5435 | Yes |
| 36 | CRABP2 | 5602 | 0.256 | 0.5500 | Yes |
| 37 | EPHX2 | 5781 | 0.244 | 0.5469 | No |
| 38 | NR1I2 | 5927 | 0.235 | 0.5454 | No |
| 39 | PEX11A | 6062 | 0.228 | 0.5443 | No |
| 40 | CEL | 6121 | 0.225 | 0.5473 | No |
| 41 | SLC25A17 | 6526 | 0.204 | 0.5305 | No |
| 42 | IDH1 | 6803 | 0.191 | 0.5204 | No |
| 43 | PABPC1 | 6917 | 0.186 | 0.5193 | No |
| 44 | RETSAT | 7043 | 0.181 | 0.5174 | No |
| 45 | ABCD2 | 7503 | 0.164 | 0.4964 | No |
| 46 | PEX5 | 7629 | 0.160 | 0.4939 | No |
| 47 | ABCC5 | 8009 | 0.147 | 0.4768 | No |
| 48 | ITGB1BP1 | 8152 | 0.142 | 0.4729 | No |
| 49 | ERCC1 | 8269 | 0.138 | 0.4703 | No |
| 50 | ACAA1 | 8344 | 0.136 | 0.4699 | No |
| 51 | ISOC1 | 8445 | 0.133 | 0.4681 | No |
| 52 | ACOX1 | 8727 | 0.126 | 0.4559 | No |
| 53 | SULT2B1 | 8855 | 0.123 | 0.4523 | No |
| 54 | PEX13 | 9010 | 0.119 | 0.4470 | No |
| 55 | SCP2 | 9105 | 0.117 | 0.4450 | No |
| 56 | FABP6 | 9258 | 0.112 | 0.4397 | No |
| 57 | GNPAT | 9321 | 0.111 | 0.4393 | No |
| 58 | UGT2B17 | 9701 | 0.103 | 0.4211 | No |
| 59 | ECI2 | 9769 | 0.101 | 0.4202 | No |
| 60 | VPS4B | 9941 | 0.098 | 0.4133 | No |
| 61 | RDH11 | 10144 | 0.094 | 0.4047 | No |
| 62 | ALDH9A1 | 10273 | 0.092 | 0.4001 | No |
| 63 | PEX11B | 10457 | 0.089 | 0.3924 | No |
| 64 | BCL10 | 10608 | 0.086 | 0.3865 | No |
| 65 | SLC25A19 | 10863 | 0.082 | 0.3746 | No |
| 66 | FDPS | 11051 | 0.078 | 0.3663 | No |
| 67 | HSD11B2 | 11343 | 0.074 | 0.3521 | No |
| 68 | DHCR24 | 11545 | 0.071 | 0.3429 | No |
| 69 | SLC23A2 | 11597 | 0.070 | 0.3420 | No |
| 70 | ACSL1 | 11679 | 0.069 | 0.3394 | No |
| 71 | SOD1 | 11745 | 0.068 | 0.3377 | No |
| 72 | SERPINA6 | 11797 | 0.067 | 0.3367 | No |
| 73 | EHHADH | 12046 | 0.063 | 0.3246 | No |
| 74 | HSD17B11 | 12326 | 0.060 | 0.3107 | No |
| 75 | SMARCC1 | 12954 | 0.051 | 0.2771 | No |
| 76 | SOD2 | 13630 | 0.042 | 0.2406 | No |
| 77 | RXRG | 13693 | 0.041 | 0.2383 | No |
| 78 | PEX14 | 13716 | 0.041 | 0.2382 | No |
| 79 | MVP | 14593 | -0.000 | 0.1892 | No |
| 80 | ACSL4 | 14707 | -0.000 | 0.1829 | No |
| 81 | SEMA3C | 14752 | -0.000 | 0.1805 | No |
| 82 | TSPO | 14905 | -0.000 | 0.1720 | No |
| 83 | STS | 14969 | -0.000 | 0.1685 | No |
| 84 | ABCD1 | 14984 | -0.000 | 0.1677 | No |
| 85 | TTR | 15385 | -0.000 | 0.1453 | No |
| 86 | IDE | 15419 | -0.000 | 0.1435 | No |
| 87 | ATXN1 | 15550 | -0.000 | 0.1362 | No |
| 88 | PRDX5 | 15600 | -0.000 | 0.1335 | No |
| 89 | YWHAH | 15629 | -0.000 | 0.1319 | No |
| 90 | DLG4 | 15742 | -0.000 | 0.1257 | No |
| 91 | SLC27A2 | 15965 | -0.000 | 0.1133 | No |
| 92 | CACNA1B | 16147 | -0.000 | 0.1032 | No |
| 93 | ABCB9 | 16179 | -0.000 | 0.1014 | No |
| 94 | CTBP1 | 16358 | -0.000 | 0.0915 | No |
| 95 | DHRS3 | 16413 | -0.000 | 0.0885 | No |
| 96 | ALDH1A1 | 16547 | -0.000 | 0.0810 | No |
| 97 | CNBP | 16757 | -0.000 | 0.0694 | No |
| 98 | CTPS1 | 16853 | -0.000 | 0.0641 | No |
| 99 | CADM1 | 17176 | -0.000 | 0.0461 | No |
| 100 | SIAH1 | 17449 | -0.000 | 0.0309 | No |
| 101 | NUDT19 | 17693 | -0.000 | 0.0173 | No |