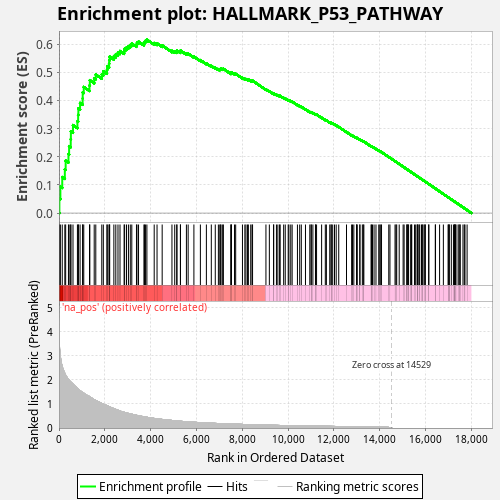
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_P53_PATHWAY |
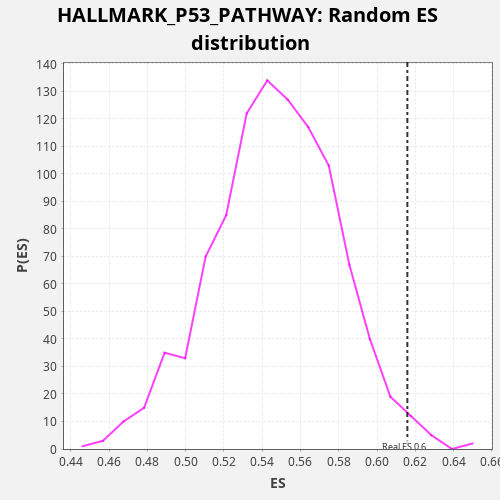
| Enrichment Score (ES) | 0.6158497 |
| Normalized Enrichment Score (NES) | 1.1263635 |
| Nominal p-value | 0.016 |
| FDR q-value | 0.25655755 |
| FWER p-Value | 0.909 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | APAF1 | 17 | 3.512 | 0.0516 | Yes |
| 2 | HSPA4L | 60 | 2.980 | 0.0938 | Yes |
| 3 | FBXW7 | 142 | 2.578 | 0.1277 | Yes |
| 4 | CD81 | 255 | 2.255 | 0.1552 | Yes |
| 5 | COQ8A | 291 | 2.193 | 0.1860 | Yes |
| 6 | RALGDS | 413 | 2.017 | 0.2094 | Yes |
| 7 | CCND3 | 442 | 1.984 | 0.2375 | Yes |
| 8 | KRT17 | 512 | 1.910 | 0.2621 | Yes |
| 9 | JAG2 | 519 | 1.902 | 0.2902 | Yes |
| 10 | PERP | 611 | 1.821 | 0.3124 | Yes |
| 11 | ABAT | 804 | 1.638 | 0.3261 | Yes |
| 12 | AEN | 836 | 1.612 | 0.3484 | Yes |
| 13 | F2R | 846 | 1.607 | 0.3720 | Yes |
| 14 | H2AJ | 920 | 1.547 | 0.3910 | Yes |
| 15 | DDIT4 | 1032 | 1.475 | 0.4068 | Yes |
| 16 | STOM | 1035 | 1.473 | 0.4287 | Yes |
| 17 | SESN1 | 1081 | 1.445 | 0.4478 | Yes |
| 18 | SDC1 | 1332 | 1.302 | 0.4533 | Yes |
| 19 | DEF6 | 1346 | 1.288 | 0.4718 | Yes |
| 20 | EI24 | 1537 | 1.176 | 0.4787 | Yes |
| 21 | RRAD | 1606 | 1.139 | 0.4919 | Yes |
| 22 | CDKN2B | 1868 | 1.019 | 0.4925 | Yes |
| 23 | DGKA | 1935 | 0.990 | 0.5036 | Yes |
| 24 | ADA | 2093 | 0.926 | 0.5086 | Yes |
| 25 | IL1A | 2106 | 0.921 | 0.5217 | Yes |
| 26 | TSPYL2 | 2187 | 0.885 | 0.5305 | Yes |
| 27 | VDR | 2193 | 0.884 | 0.5434 | Yes |
| 28 | SLC35D1 | 2213 | 0.875 | 0.5554 | Yes |
| 29 | DNTTIP2 | 2395 | 0.807 | 0.5573 | Yes |
| 30 | DDB2 | 2480 | 0.772 | 0.5641 | Yes |
| 31 | FUCA1 | 2575 | 0.736 | 0.5699 | Yes |
| 32 | FAM162A | 2663 | 0.704 | 0.5755 | Yes |
| 33 | MKNK2 | 2842 | 0.651 | 0.5752 | Yes |
| 34 | RAD9A | 2865 | 0.643 | 0.5836 | Yes |
| 35 | NUDT15 | 2947 | 0.624 | 0.5884 | Yes |
| 36 | GPX2 | 3031 | 0.604 | 0.5928 | Yes |
| 37 | PIDD1 | 3112 | 0.585 | 0.5970 | Yes |
| 38 | PMM1 | 3178 | 0.571 | 0.6019 | Yes |
| 39 | EPS8L2 | 3382 | 0.527 | 0.5984 | Yes |
| 40 | IRAG2 | 3404 | 0.522 | 0.6050 | Yes |
| 41 | CD82 | 3472 | 0.509 | 0.6089 | Yes |
| 42 | TXNIP | 3708 | 0.464 | 0.6026 | Yes |
| 43 | STEAP3 | 3734 | 0.459 | 0.6081 | Yes |
| 44 | TRIB3 | 3779 | 0.453 | 0.6124 | Yes |
| 45 | XPC | 3837 | 0.444 | 0.6158 | Yes |
| 46 | UPP1 | 4156 | 0.396 | 0.6039 | No |
| 47 | CCP110 | 4280 | 0.380 | 0.6027 | No |
| 48 | TM7SF3 | 4504 | 0.354 | 0.5955 | No |
| 49 | ZNF365 | 4931 | 0.312 | 0.5762 | No |
| 50 | ANKRA2 | 5052 | 0.301 | 0.5740 | No |
| 51 | PPP1R15A | 5141 | 0.293 | 0.5734 | No |
| 52 | HRAS | 5149 | 0.293 | 0.5774 | No |
| 53 | NUPR1 | 5300 | 0.280 | 0.5732 | No |
| 54 | MAPKAPK3 | 5303 | 0.279 | 0.5773 | No |
| 55 | GLS2 | 5562 | 0.259 | 0.5666 | No |
| 56 | FDXR | 5638 | 0.253 | 0.5662 | No |
| 57 | RRP8 | 5889 | 0.237 | 0.5557 | No |
| 58 | RAD51C | 6172 | 0.222 | 0.5432 | No |
| 59 | RNF19B | 6434 | 0.209 | 0.5317 | No |
| 60 | SLC7A11 | 6649 | 0.198 | 0.5226 | No |
| 61 | PLXNB2 | 6822 | 0.190 | 0.5158 | No |
| 62 | EPHA2 | 6958 | 0.185 | 0.5110 | No |
| 63 | PLK3 | 7022 | 0.181 | 0.5102 | No |
| 64 | RETSAT | 7043 | 0.181 | 0.5118 | No |
| 65 | RCHY1 | 7074 | 0.179 | 0.5128 | No |
| 66 | SERPINB5 | 7145 | 0.176 | 0.5115 | No |
| 67 | TAP1 | 7176 | 0.175 | 0.5124 | No |
| 68 | NDRG1 | 7505 | 0.164 | 0.4964 | No |
| 69 | ELP1 | 7509 | 0.164 | 0.4987 | No |
| 70 | ITGB4 | 7530 | 0.163 | 0.5000 | No |
| 71 | BAK1 | 7663 | 0.158 | 0.4950 | No |
| 72 | APP | 7713 | 0.157 | 0.4946 | No |
| 73 | ABCC5 | 8009 | 0.147 | 0.4802 | No |
| 74 | PTPN14 | 8114 | 0.144 | 0.4765 | No |
| 75 | HMOX1 | 8162 | 0.142 | 0.4760 | No |
| 76 | WRAP73 | 8231 | 0.140 | 0.4743 | No |
| 77 | RPL36 | 8267 | 0.138 | 0.4744 | No |
| 78 | SERTAD3 | 8370 | 0.135 | 0.4707 | No |
| 79 | TRAFD1 | 8427 | 0.134 | 0.4695 | No |
| 80 | WWP1 | 8450 | 0.133 | 0.4703 | No |
| 81 | TM4SF1 | 9034 | 0.118 | 0.4393 | No |
| 82 | CDKN1A | 9183 | 0.114 | 0.4327 | No |
| 83 | PRKAB1 | 9366 | 0.110 | 0.4242 | No |
| 84 | RPL18 | 9373 | 0.110 | 0.4255 | No |
| 85 | TPD52L1 | 9493 | 0.107 | 0.4204 | No |
| 86 | RPS27L | 9554 | 0.106 | 0.4186 | No |
| 87 | POM121 | 9631 | 0.104 | 0.4159 | No |
| 88 | PROCR | 9654 | 0.104 | 0.4162 | No |
| 89 | SPHK1 | 9808 | 0.100 | 0.4091 | No |
| 90 | TCN2 | 9877 | 0.099 | 0.4068 | No |
| 91 | ALOX15B | 10006 | 0.097 | 0.4011 | No |
| 92 | RHBDF2 | 10037 | 0.096 | 0.4008 | No |
| 93 | DCXR | 10118 | 0.095 | 0.3978 | No |
| 94 | ST14 | 10178 | 0.094 | 0.3958 | No |
| 95 | ABHD4 | 10409 | 0.090 | 0.3843 | No |
| 96 | PPM1D | 10508 | 0.088 | 0.3801 | No |
| 97 | BTG2 | 10578 | 0.087 | 0.3775 | No |
| 98 | BAIAP2 | 10760 | 0.084 | 0.3686 | No |
| 99 | CGRRF1 | 10949 | 0.080 | 0.3593 | No |
| 100 | POLH | 11006 | 0.079 | 0.3573 | No |
| 101 | TPRKB | 11013 | 0.079 | 0.3581 | No |
| 102 | SLC19A2 | 11084 | 0.078 | 0.3554 | No |
| 103 | CDKN2AIP | 11196 | 0.076 | 0.3503 | No |
| 104 | CASP1 | 11202 | 0.076 | 0.3511 | No |
| 105 | CCNG1 | 11244 | 0.076 | 0.3500 | No |
| 106 | OSGIN1 | 11465 | 0.072 | 0.3387 | No |
| 107 | CDH13 | 11626 | 0.070 | 0.3307 | No |
| 108 | FGF13 | 11673 | 0.069 | 0.3292 | No |
| 109 | CTSF | 11820 | 0.067 | 0.3220 | No |
| 110 | KIF13B | 11879 | 0.066 | 0.3197 | No |
| 111 | ISCU | 11883 | 0.066 | 0.3205 | No |
| 112 | CYFIP2 | 11948 | 0.064 | 0.3179 | No |
| 113 | PRMT2 | 12027 | 0.063 | 0.3145 | No |
| 114 | BAX | 12101 | 0.063 | 0.3113 | No |
| 115 | RAB40C | 12212 | 0.061 | 0.3060 | No |
| 116 | ZMAT3 | 12551 | 0.056 | 0.2879 | No |
| 117 | AK1 | 12775 | 0.053 | 0.2762 | No |
| 118 | ATF3 | 12813 | 0.053 | 0.2749 | No |
| 119 | TGFA | 12871 | 0.052 | 0.2725 | No |
| 120 | DDIT3 | 12990 | 0.051 | 0.2666 | No |
| 121 | H1-2 | 12996 | 0.051 | 0.2671 | No |
| 122 | HINT1 | 13019 | 0.050 | 0.2666 | No |
| 123 | LDHB | 13133 | 0.049 | 0.2610 | No |
| 124 | VAMP8 | 13137 | 0.048 | 0.2615 | No |
| 125 | PITPNC1 | 13243 | 0.047 | 0.2564 | No |
| 126 | HDAC3 | 13285 | 0.047 | 0.2548 | No |
| 127 | TRIAP1 | 13309 | 0.046 | 0.2542 | No |
| 128 | DRAM1 | 13623 | 0.042 | 0.2372 | No |
| 129 | ACVR1B | 13644 | 0.042 | 0.2367 | No |
| 130 | NOL8 | 13686 | 0.041 | 0.2350 | No |
| 131 | TRAF4 | 13754 | 0.040 | 0.2319 | No |
| 132 | RGS16 | 13838 | 0.039 | 0.2278 | No |
| 133 | SEC61A1 | 13959 | 0.037 | 0.2216 | No |
| 134 | CLCA2 | 14004 | 0.036 | 0.2197 | No |
| 135 | EPHX1 | 14047 | 0.035 | 0.2178 | No |
| 136 | PLK2 | 14080 | 0.035 | 0.2166 | No |
| 137 | BTG1 | 14396 | 0.022 | 0.1992 | No |
| 138 | BMP2 | 14448 | 0.020 | 0.1966 | No |
| 139 | MXD1 | 14670 | -0.000 | 0.1842 | No |
| 140 | IP6K2 | 14711 | -0.000 | 0.1820 | No |
| 141 | TP63 | 14743 | -0.000 | 0.1803 | No |
| 142 | CCNK | 14852 | -0.000 | 0.1742 | No |
| 143 | TSC22D1 | 15023 | -0.000 | 0.1646 | No |
| 144 | TGFB1 | 15066 | -0.000 | 0.1623 | No |
| 145 | ZBTB16 | 15188 | -0.000 | 0.1555 | No |
| 146 | VWA5A | 15190 | -0.000 | 0.1554 | No |
| 147 | CSRNP2 | 15206 | -0.000 | 0.1546 | No |
| 148 | RPS12 | 15243 | -0.000 | 0.1526 | No |
| 149 | HBEGF | 15262 | -0.000 | 0.1516 | No |
| 150 | GADD45A | 15351 | -0.000 | 0.1466 | No |
| 151 | FOXO3 | 15393 | -0.000 | 0.1443 | No |
| 152 | CCND2 | 15397 | -0.000 | 0.1442 | No |
| 153 | MXD4 | 15521 | -0.000 | 0.1373 | No |
| 154 | TNFSF9 | 15571 | -0.000 | 0.1345 | No |
| 155 | LIF | 15635 | -0.000 | 0.1310 | No |
| 156 | KLK8 | 15655 | -0.000 | 0.1299 | No |
| 157 | SAT1 | 15668 | -0.000 | 0.1292 | No |
| 158 | NINJ1 | 15718 | -0.000 | 0.1265 | No |
| 159 | PCNA | 15747 | -0.000 | 0.1249 | No |
| 160 | MDM2 | 15825 | -0.000 | 0.1206 | No |
| 161 | KLF4 | 15860 | -0.000 | 0.1187 | No |
| 162 | IER3 | 15886 | -0.000 | 0.1173 | No |
| 163 | RB1 | 15945 | -0.000 | 0.1140 | No |
| 164 | TOB1 | 15983 | -0.000 | 0.1119 | No |
| 165 | TP53 | 15990 | -0.000 | 0.1116 | No |
| 166 | CDKN2A | 16138 | -0.000 | 0.1033 | No |
| 167 | NOTCH1 | 16146 | -0.000 | 0.1030 | No |
| 168 | IER5 | 16427 | -0.000 | 0.0872 | No |
| 169 | INHBB | 16437 | -0.000 | 0.0867 | No |
| 170 | BLCAP | 16615 | -0.000 | 0.0768 | No |
| 171 | FOS | 16780 | -0.000 | 0.0676 | No |
| 172 | SFN | 16986 | -0.000 | 0.0561 | No |
| 173 | CDK5R1 | 17012 | -0.000 | 0.0547 | No |
| 174 | NHLH2 | 17032 | -0.000 | 0.0536 | No |
| 175 | JUN | 17035 | -0.000 | 0.0535 | No |
| 176 | RAP2B | 17127 | -0.000 | 0.0484 | No |
| 177 | IRAK1 | 17220 | -0.000 | 0.0432 | No |
| 178 | SOCS1 | 17268 | -0.000 | 0.0406 | No |
| 179 | SP1 | 17276 | -0.000 | 0.0402 | No |
| 180 | ZFP36L1 | 17280 | -0.000 | 0.0400 | No |
| 181 | RXRA | 17312 | -0.000 | 0.0383 | No |
| 182 | HEXIM1 | 17331 | -0.000 | 0.0373 | No |
| 183 | S100A4 | 17423 | -0.000 | 0.0322 | No |
| 184 | GM2A | 17465 | -0.000 | 0.0299 | No |
| 185 | PDGFA | 17504 | -0.000 | 0.0277 | No |
| 186 | S100A10 | 17513 | -0.000 | 0.0273 | No |
| 187 | RACK1 | 17636 | -0.000 | 0.0204 | No |
| 188 | TAX1BP3 | 17695 | -0.000 | 0.0172 | No |
| 189 | IFI30 | 17723 | -0.000 | 0.0157 | No |
| 190 | CEBPA | 17816 | -0.000 | 0.0105 | No |