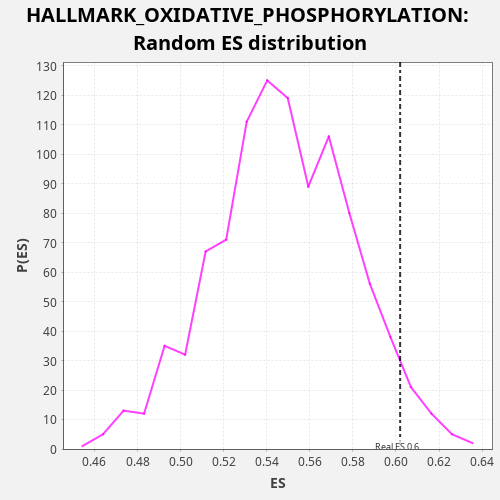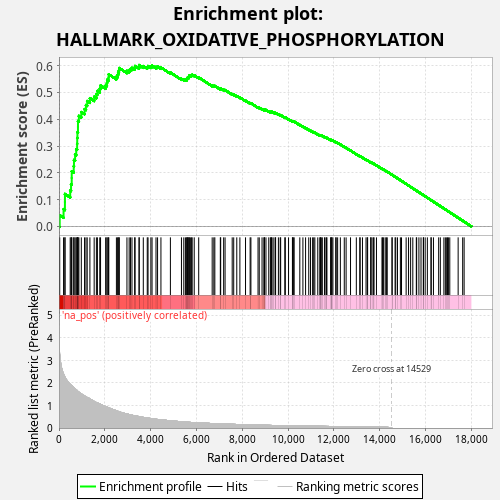
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_OXIDATIVE_PHOSPHORYLATION |
| Enrichment Score (ES) | 0.6019309 |
| Normalized Enrichment Score (NES) | 1.0993003 |
| Nominal p-value | 0.04 |
| FDR q-value | 0.34069303 |
| FWER p-Value | 0.994 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | COX7A2 | 31 | 3.268 | 0.0418 | Yes |
| 2 | ETFDH | 199 | 2.396 | 0.0644 | Yes |
| 3 | PDHA1 | 259 | 2.251 | 0.0911 | Yes |
| 4 | SLC25A4 | 260 | 2.250 | 0.1211 | Yes |
| 5 | GOT2 | 489 | 1.929 | 0.1340 | Yes |
| 6 | SLC25A5 | 526 | 1.898 | 0.1573 | Yes |
| 7 | ATP5F1C | 555 | 1.876 | 0.1807 | Yes |
| 8 | NDUFC2 | 556 | 1.874 | 0.2057 | Yes |
| 9 | HTRA2 | 643 | 1.786 | 0.2247 | Yes |
| 10 | TIMM9 | 654 | 1.780 | 0.2479 | Yes |
| 11 | PMPCA | 703 | 1.735 | 0.2683 | Yes |
| 12 | NDUFC1 | 752 | 1.696 | 0.2882 | Yes |
| 13 | PDP1 | 792 | 1.654 | 0.3081 | Yes |
| 14 | NDUFB7 | 796 | 1.648 | 0.3299 | Yes |
| 15 | BCKDHA | 805 | 1.637 | 0.3513 | Yes |
| 16 | UQCR11 | 828 | 1.620 | 0.3716 | Yes |
| 17 | ATP5F1D | 829 | 1.619 | 0.3932 | Yes |
| 18 | ATP5PB | 868 | 1.589 | 0.4123 | Yes |
| 19 | NDUFB1 | 974 | 1.507 | 0.4265 | Yes |
| 20 | MTRR | 1115 | 1.420 | 0.4376 | Yes |
| 21 | TIMM13 | 1177 | 1.385 | 0.4526 | Yes |
| 22 | IDH2 | 1231 | 1.354 | 0.4677 | Yes |
| 23 | CYC1 | 1349 | 1.288 | 0.4783 | Yes |
| 24 | ATP6V1G1 | 1535 | 1.177 | 0.4836 | Yes |
| 25 | OPA1 | 1632 | 1.122 | 0.4931 | Yes |
| 26 | ALAS1 | 1677 | 1.108 | 0.5054 | Yes |
| 27 | NNT | 1780 | 1.062 | 0.5139 | Yes |
| 28 | UQCRC2 | 1814 | 1.048 | 0.5260 | Yes |
| 29 | NDUFA5 | 2036 | 0.949 | 0.5262 | Yes |
| 30 | NDUFB2 | 2091 | 0.926 | 0.5356 | Yes |
| 31 | MRPS15 | 2104 | 0.921 | 0.5472 | Yes |
| 32 | FH | 2167 | 0.894 | 0.5556 | Yes |
| 33 | DLD | 2169 | 0.894 | 0.5675 | Yes |
| 34 | ATP6V1C1 | 2506 | 0.763 | 0.5588 | Yes |
| 35 | DLAT | 2560 | 0.743 | 0.5657 | Yes |
| 36 | UQCRC1 | 2584 | 0.733 | 0.5742 | Yes |
| 37 | ATP5MF | 2615 | 0.723 | 0.5821 | Yes |
| 38 | PHB2 | 2631 | 0.714 | 0.5908 | Yes |
| 39 | MRPS30 | 2966 | 0.619 | 0.5803 | Yes |
| 40 | PRDX3 | 3063 | 0.596 | 0.5829 | Yes |
| 41 | SURF1 | 3120 | 0.583 | 0.5875 | Yes |
| 42 | SUCLG1 | 3186 | 0.570 | 0.5915 | Yes |
| 43 | NDUFS3 | 3303 | 0.542 | 0.5922 | Yes |
| 44 | OXA1L | 3321 | 0.539 | 0.5984 | Yes |
| 45 | RHOT2 | 3497 | 0.503 | 0.5953 | Yes |
| 46 | BDH2 | 3499 | 0.502 | 0.6019 | Yes |
| 47 | ETFA | 3680 | 0.469 | 0.5981 | No |
| 48 | NDUFV2 | 3853 | 0.442 | 0.5943 | No |
| 49 | MDH2 | 3887 | 0.435 | 0.5983 | No |
| 50 | COX15 | 4010 | 0.420 | 0.5970 | No |
| 51 | PDK4 | 4062 | 0.412 | 0.5996 | No |
| 52 | RHOT1 | 4235 | 0.386 | 0.5951 | No |
| 53 | PHYH | 4303 | 0.377 | 0.5964 | No |
| 54 | NDUFS4 | 4450 | 0.361 | 0.5930 | No |
| 55 | NDUFA6 | 4863 | 0.319 | 0.5741 | No |
| 56 | ACADM | 5346 | 0.275 | 0.5507 | No |
| 57 | ACAA2 | 5448 | 0.267 | 0.5486 | No |
| 58 | NDUFV1 | 5519 | 0.261 | 0.5482 | No |
| 59 | ACAT1 | 5581 | 0.258 | 0.5482 | No |
| 60 | MRPL15 | 5584 | 0.258 | 0.5515 | No |
| 61 | HADHA | 5600 | 0.257 | 0.5541 | No |
| 62 | COX6A1 | 5627 | 0.254 | 0.5560 | No |
| 63 | LDHA | 5673 | 0.251 | 0.5568 | No |
| 64 | NDUFB3 | 5675 | 0.251 | 0.5601 | No |
| 65 | SLC25A20 | 5685 | 0.250 | 0.5630 | No |
| 66 | NQO2 | 5746 | 0.246 | 0.5629 | No |
| 67 | MRPL35 | 5790 | 0.243 | 0.5637 | No |
| 68 | PDHX | 5812 | 0.242 | 0.5657 | No |
| 69 | DECR1 | 5903 | 0.236 | 0.5638 | No |
| 70 | HADHB | 6099 | 0.226 | 0.5559 | No |
| 71 | ATP5PD | 6687 | 0.196 | 0.5256 | No |
| 72 | TIMM8B | 6759 | 0.193 | 0.5241 | No |
| 73 | IDH1 | 6803 | 0.191 | 0.5243 | No |
| 74 | RETSAT | 7043 | 0.181 | 0.5133 | No |
| 75 | NDUFA4 | 7058 | 0.180 | 0.5149 | No |
| 76 | ECHS1 | 7188 | 0.175 | 0.5100 | No |
| 77 | MRPL34 | 7250 | 0.173 | 0.5088 | No |
| 78 | CPT1A | 7561 | 0.162 | 0.4936 | No |
| 79 | GPI | 7627 | 0.160 | 0.4921 | No |
| 80 | TIMM50 | 7761 | 0.155 | 0.4867 | No |
| 81 | NDUFA9 | 7897 | 0.151 | 0.4811 | No |
| 82 | NDUFB4 | 8143 | 0.143 | 0.4693 | No |
| 83 | ACAA1 | 8344 | 0.136 | 0.4598 | No |
| 84 | ALDH6A1 | 8378 | 0.135 | 0.4598 | No |
| 85 | MRPL11 | 8689 | 0.127 | 0.4441 | No |
| 86 | ATP6V1D | 8742 | 0.126 | 0.4428 | No |
| 87 | ATP5PO | 8864 | 0.123 | 0.4377 | No |
| 88 | ATP6V1H | 8947 | 0.120 | 0.4347 | No |
| 89 | SUCLA2 | 8960 | 0.120 | 0.4356 | No |
| 90 | UQCRB | 8972 | 0.120 | 0.4366 | No |
| 91 | OAT | 9035 | 0.118 | 0.4347 | No |
| 92 | TIMM10 | 9157 | 0.115 | 0.4294 | No |
| 93 | MTX2 | 9228 | 0.113 | 0.4270 | No |
| 94 | GPX4 | 9255 | 0.112 | 0.4270 | No |
| 95 | CYB5R3 | 9278 | 0.112 | 0.4273 | No |
| 96 | SUPV3L1 | 9303 | 0.111 | 0.4274 | No |
| 97 | IMMT | 9380 | 0.109 | 0.4246 | No |
| 98 | ATP5MC3 | 9444 | 0.108 | 0.4225 | No |
| 99 | UQCR10 | 9456 | 0.108 | 0.4234 | No |
| 100 | NDUFB6 | 9578 | 0.106 | 0.4180 | No |
| 101 | NDUFS8 | 9656 | 0.104 | 0.4150 | No |
| 102 | ACADVL | 9677 | 0.104 | 0.4153 | No |
| 103 | NDUFB8 | 9859 | 0.099 | 0.4065 | No |
| 104 | CS | 9879 | 0.099 | 0.4067 | No |
| 105 | MDH1 | 10021 | 0.097 | 0.4001 | No |
| 106 | FXN | 10190 | 0.093 | 0.3919 | No |
| 107 | POLR2F | 10200 | 0.093 | 0.3926 | No |
| 108 | FDX1 | 10231 | 0.093 | 0.3922 | No |
| 109 | COX11 | 10266 | 0.092 | 0.3915 | No |
| 110 | NDUFS7 | 10513 | 0.088 | 0.3789 | No |
| 111 | POR | 10648 | 0.086 | 0.3725 | No |
| 112 | TCIRG1 | 10765 | 0.084 | 0.3671 | No |
| 113 | IDH3A | 10903 | 0.081 | 0.3605 | No |
| 114 | UQCRFS1 | 10981 | 0.080 | 0.3572 | No |
| 115 | SLC25A12 | 11072 | 0.078 | 0.3532 | No |
| 116 | SDHB | 11120 | 0.077 | 0.3516 | No |
| 117 | MRPS11 | 11169 | 0.077 | 0.3499 | No |
| 118 | MGST3 | 11297 | 0.075 | 0.3438 | No |
| 119 | OGDH | 11388 | 0.073 | 0.3397 | No |
| 120 | ATP6V1F | 11407 | 0.073 | 0.3397 | No |
| 121 | ATP6V0E1 | 11418 | 0.073 | 0.3401 | No |
| 122 | SDHC | 11452 | 0.072 | 0.3392 | No |
| 123 | NDUFS2 | 11495 | 0.072 | 0.3378 | No |
| 124 | PDHB | 11589 | 0.070 | 0.3335 | No |
| 125 | ETFB | 11616 | 0.070 | 0.3330 | No |
| 126 | DLST | 11680 | 0.069 | 0.3304 | No |
| 127 | NDUFA8 | 11696 | 0.068 | 0.3304 | No |
| 128 | NDUFB5 | 11858 | 0.066 | 0.3223 | No |
| 129 | ISCU | 11883 | 0.066 | 0.3218 | No |
| 130 | NDUFS1 | 11884 | 0.066 | 0.3227 | No |
| 131 | SLC25A3 | 11916 | 0.065 | 0.3218 | No |
| 132 | ACO2 | 11958 | 0.064 | 0.3204 | No |
| 133 | SDHA | 12080 | 0.063 | 0.3144 | No |
| 134 | BAX | 12101 | 0.063 | 0.3141 | No |
| 135 | GLUD1 | 12164 | 0.062 | 0.3115 | No |
| 136 | COX6C | 12283 | 0.060 | 0.3056 | No |
| 137 | ATP5ME | 12454 | 0.058 | 0.2969 | No |
| 138 | COX10 | 12534 | 0.057 | 0.2932 | No |
| 139 | ATP5MC2 | 12727 | 0.054 | 0.2831 | No |
| 140 | COX5B | 12980 | 0.051 | 0.2696 | No |
| 141 | LDHB | 13133 | 0.049 | 0.2618 | No |
| 142 | ATP6V0B | 13145 | 0.048 | 0.2618 | No |
| 143 | ATP6V1E1 | 13248 | 0.047 | 0.2567 | No |
| 144 | ATP5F1E | 13404 | 0.045 | 0.2486 | No |
| 145 | MAOB | 13460 | 0.044 | 0.2461 | No |
| 146 | UQCRQ | 13474 | 0.044 | 0.2460 | No |
| 147 | CYB5A | 13609 | 0.042 | 0.2390 | No |
| 148 | GRPEL1 | 13615 | 0.042 | 0.2393 | No |
| 149 | ATP5F1A | 13666 | 0.041 | 0.2370 | No |
| 150 | COX7A2L | 13735 | 0.040 | 0.2337 | No |
| 151 | CASP7 | 13751 | 0.040 | 0.2334 | No |
| 152 | SLC25A6 | 13859 | 0.039 | 0.2279 | No |
| 153 | ATP1B1 | 14100 | 0.034 | 0.2149 | No |
| 154 | ABCB7 | 14126 | 0.034 | 0.2140 | No |
| 155 | COX4I1 | 14171 | 0.033 | 0.2119 | No |
| 156 | ATP5F1B | 14241 | 0.031 | 0.2085 | No |
| 157 | NDUFA1 | 14293 | 0.029 | 0.2060 | No |
| 158 | ATP6AP1 | 14329 | 0.027 | 0.2044 | No |
| 159 | NDUFAB1 | 14539 | -0.000 | 0.1927 | No |
| 160 | HCCS | 14543 | -0.000 | 0.1925 | No |
| 161 | MPC1 | 14672 | -0.000 | 0.1853 | No |
| 162 | IDH3G | 14701 | -0.000 | 0.1837 | No |
| 163 | VDAC3 | 14776 | -0.000 | 0.1796 | No |
| 164 | TOMM22 | 14903 | -0.000 | 0.1725 | No |
| 165 | IDH3B | 14953 | -0.000 | 0.1698 | No |
| 166 | SLC25A11 | 15155 | -0.000 | 0.1585 | No |
| 167 | HSPA9 | 15260 | -0.000 | 0.1526 | No |
| 168 | MFN2 | 15350 | -0.000 | 0.1476 | No |
| 169 | MTRF1 | 15446 | -0.000 | 0.1423 | No |
| 170 | COX6B1 | 15599 | -0.000 | 0.1338 | No |
| 171 | COX7C | 15614 | -0.000 | 0.1330 | No |
| 172 | NDUFA2 | 15712 | -0.000 | 0.1275 | No |
| 173 | ISCA1 | 15804 | -0.000 | 0.1224 | No |
| 174 | LRPPRC | 15904 | -0.000 | 0.1169 | No |
| 175 | COX17 | 15912 | -0.000 | 0.1165 | No |
| 176 | AFG3L2 | 15986 | -0.000 | 0.1124 | No |
| 177 | NDUFS6 | 16081 | -0.000 | 0.1071 | No |
| 178 | TOMM70 | 16241 | -0.000 | 0.0982 | No |
| 179 | ATP5PF | 16248 | -0.000 | 0.0978 | No |
| 180 | ATP5MC1 | 16347 | -0.000 | 0.0923 | No |
| 181 | VDAC2 | 16575 | -0.000 | 0.0796 | No |
| 182 | ATP5MG | 16648 | -0.000 | 0.0756 | No |
| 183 | NDUFA3 | 16803 | -0.000 | 0.0669 | No |
| 184 | CYCS | 16867 | -0.000 | 0.0634 | No |
| 185 | UQCRH | 16925 | -0.000 | 0.0602 | No |
| 186 | MRPS22 | 16962 | -0.000 | 0.0582 | No |
| 187 | COX8A | 16999 | -0.000 | 0.0561 | No |
| 188 | COX5A | 17060 | -0.000 | 0.0528 | No |
| 189 | ACADSB | 17425 | -0.000 | 0.0323 | No |
| 190 | SDHD | 17623 | -0.000 | 0.0213 | No |
| 191 | VDAC1 | 17686 | -0.000 | 0.0178 | No |