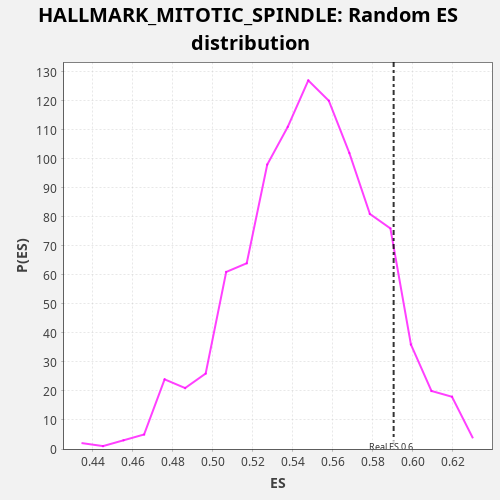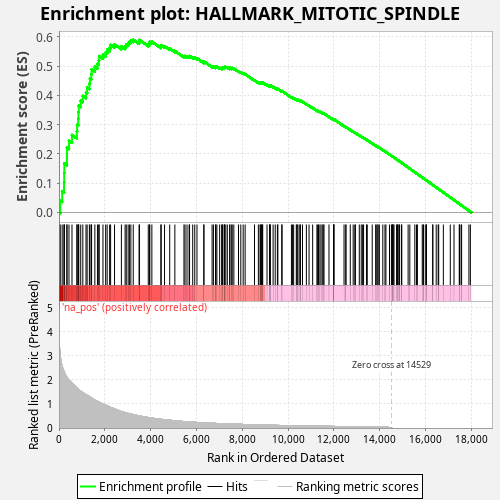
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_MITOTIC_SPINDLE |
| Enrichment Score (ES) | 0.5904487 |
| Normalized Enrichment Score (NES) | 1.0765252 |
| Nominal p-value | 0.108 |
| FDR q-value | 0.45856285 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | CTTN | 55 | 3.025 | 0.0403 | Yes |
| 2 | MAP3K11 | 138 | 2.582 | 0.0727 | Yes |
| 3 | RFC1 | 220 | 2.338 | 0.1017 | Yes |
| 4 | TTK | 226 | 2.324 | 0.1348 | Yes |
| 5 | EZR | 233 | 2.309 | 0.1675 | Yes |
| 6 | ARHGAP4 | 346 | 2.107 | 0.1915 | Yes |
| 7 | ARHGAP5 | 347 | 2.101 | 0.2216 | Yes |
| 8 | ARHGEF2 | 436 | 1.991 | 0.2452 | Yes |
| 9 | NF1 | 567 | 1.864 | 0.2647 | Yes |
| 10 | HOOK3 | 778 | 1.671 | 0.2768 | Yes |
| 11 | SMC3 | 793 | 1.654 | 0.2998 | Yes |
| 12 | DOCK4 | 842 | 1.610 | 0.3202 | Yes |
| 13 | TUBA4A | 849 | 1.603 | 0.3428 | Yes |
| 14 | CEP57 | 863 | 1.591 | 0.3649 | Yes |
| 15 | DST | 948 | 1.531 | 0.3822 | Yes |
| 16 | RAPGEF5 | 1040 | 1.469 | 0.3981 | Yes |
| 17 | APC | 1179 | 1.383 | 0.4102 | Yes |
| 18 | ARHGAP27 | 1228 | 1.355 | 0.4269 | Yes |
| 19 | SPTAN1 | 1326 | 1.306 | 0.4402 | Yes |
| 20 | CENPE | 1352 | 1.284 | 0.4573 | Yes |
| 21 | BCR | 1412 | 1.254 | 0.4719 | Yes |
| 22 | RASA2 | 1423 | 1.250 | 0.4893 | Yes |
| 23 | RACGAP1 | 1565 | 1.163 | 0.4981 | Yes |
| 24 | TUBGCP3 | 1679 | 1.107 | 0.5076 | Yes |
| 25 | GSN | 1735 | 1.080 | 0.5200 | Yes |
| 26 | PDLIM5 | 1754 | 1.073 | 0.5344 | Yes |
| 27 | RHOF | 1920 | 0.996 | 0.5394 | Yes |
| 28 | FGD6 | 2037 | 0.948 | 0.5465 | Yes |
| 29 | BCAR1 | 2102 | 0.922 | 0.5561 | Yes |
| 30 | PKD2 | 2210 | 0.877 | 0.5627 | Yes |
| 31 | KIF5B | 2253 | 0.858 | 0.5727 | Yes |
| 32 | PCM1 | 2424 | 0.792 | 0.5745 | Yes |
| 33 | SORBS2 | 2724 | 0.683 | 0.5675 | Yes |
| 34 | SMC4 | 2886 | 0.640 | 0.5676 | Yes |
| 35 | CEP131 | 2936 | 0.628 | 0.5739 | Yes |
| 36 | CEP72 | 3018 | 0.607 | 0.5781 | Yes |
| 37 | CLIP1 | 3073 | 0.593 | 0.5835 | Yes |
| 38 | ALS2 | 3137 | 0.578 | 0.5883 | Yes |
| 39 | CNTRL | 3241 | 0.554 | 0.5904 | Yes |
| 40 | RHOT2 | 3497 | 0.503 | 0.5833 | No |
| 41 | FARP1 | 3511 | 0.501 | 0.5898 | No |
| 42 | CEP250 | 3897 | 0.434 | 0.5744 | No |
| 43 | AURKA | 3956 | 0.426 | 0.5773 | No |
| 44 | ARHGAP10 | 3962 | 0.424 | 0.5831 | No |
| 45 | CNTROB | 4054 | 0.413 | 0.5839 | No |
| 46 | SHROOM2 | 4447 | 0.361 | 0.5671 | No |
| 47 | BRCA2 | 4461 | 0.360 | 0.5715 | No |
| 48 | AKAP13 | 4607 | 0.345 | 0.5683 | No |
| 49 | KPTN | 4833 | 0.322 | 0.5603 | No |
| 50 | CDK5RAP2 | 5058 | 0.300 | 0.5520 | No |
| 51 | SOS1 | 5455 | 0.267 | 0.5336 | No |
| 52 | TBCD | 5502 | 0.263 | 0.5348 | No |
| 53 | TOP2A | 5588 | 0.258 | 0.5337 | No |
| 54 | ECT2 | 5680 | 0.251 | 0.5322 | No |
| 55 | FGD4 | 5699 | 0.249 | 0.5348 | No |
| 56 | KIF22 | 5837 | 0.240 | 0.5305 | No |
| 57 | ARAP3 | 5926 | 0.235 | 0.5289 | No |
| 58 | TAOK2 | 6023 | 0.230 | 0.5269 | No |
| 59 | GEMIN4 | 6323 | 0.214 | 0.5131 | No |
| 60 | DLGAP5 | 6326 | 0.214 | 0.5161 | No |
| 61 | RALBP1 | 6683 | 0.196 | 0.4989 | No |
| 62 | CCNB2 | 6741 | 0.193 | 0.4985 | No |
| 63 | EPB41L2 | 6833 | 0.189 | 0.4961 | No |
| 64 | SUN2 | 6847 | 0.189 | 0.4981 | No |
| 65 | MAP1S | 6893 | 0.187 | 0.4982 | No |
| 66 | BUB1 | 7013 | 0.182 | 0.4942 | No |
| 67 | CSNK1D | 7105 | 0.178 | 0.4916 | No |
| 68 | RAPGEF6 | 7119 | 0.178 | 0.4934 | No |
| 69 | NEK2 | 7132 | 0.177 | 0.4953 | No |
| 70 | TUBGCP2 | 7213 | 0.174 | 0.4933 | No |
| 71 | KIF15 | 7216 | 0.174 | 0.4957 | No |
| 72 | ANLN | 7223 | 0.174 | 0.4978 | No |
| 73 | MYO1E | 7281 | 0.171 | 0.4971 | No |
| 74 | TUBGCP6 | 7347 | 0.169 | 0.4959 | No |
| 75 | BIRC5 | 7452 | 0.165 | 0.4924 | No |
| 76 | PIF1 | 7462 | 0.165 | 0.4943 | No |
| 77 | TUBD1 | 7541 | 0.162 | 0.4922 | No |
| 78 | KATNA1 | 7559 | 0.162 | 0.4936 | No |
| 79 | ALMS1 | 7633 | 0.159 | 0.4918 | No |
| 80 | ARHGEF12 | 7836 | 0.153 | 0.4826 | No |
| 81 | KIF2C | 7937 | 0.149 | 0.4792 | No |
| 82 | LLGL1 | 8040 | 0.146 | 0.4755 | No |
| 83 | BIN1 | 8126 | 0.143 | 0.4728 | No |
| 84 | ESPL1 | 8535 | 0.131 | 0.4518 | No |
| 85 | NET1 | 8704 | 0.127 | 0.4442 | No |
| 86 | PLEKHG2 | 8784 | 0.125 | 0.4415 | No |
| 87 | KIF1B | 8785 | 0.125 | 0.4433 | No |
| 88 | CD2AP | 8828 | 0.124 | 0.4427 | No |
| 89 | TRIO | 8835 | 0.123 | 0.4442 | No |
| 90 | ITSN1 | 8880 | 0.122 | 0.4434 | No |
| 91 | SASS6 | 8893 | 0.122 | 0.4445 | No |
| 92 | MYO9B | 9081 | 0.117 | 0.4357 | No |
| 93 | CDC42EP2 | 9196 | 0.114 | 0.4309 | No |
| 94 | TIAM1 | 9204 | 0.114 | 0.4322 | No |
| 95 | ATG4B | 9215 | 0.114 | 0.4332 | No |
| 96 | KNTC1 | 9221 | 0.113 | 0.4346 | No |
| 97 | RASAL2 | 9347 | 0.110 | 0.4292 | No |
| 98 | KIF20B | 9439 | 0.108 | 0.4256 | No |
| 99 | ARFGEF1 | 9534 | 0.107 | 0.4219 | No |
| 100 | NDC80 | 9555 | 0.106 | 0.4223 | No |
| 101 | SSH2 | 9728 | 0.102 | 0.4141 | No |
| 102 | CLIP2 | 9736 | 0.102 | 0.4151 | No |
| 103 | NUSAP1 | 10147 | 0.094 | 0.3935 | No |
| 104 | KATNB1 | 10196 | 0.093 | 0.3921 | No |
| 105 | ARHGAP29 | 10244 | 0.093 | 0.3908 | No |
| 106 | SAC3D1 | 10359 | 0.091 | 0.3857 | No |
| 107 | MYH10 | 10390 | 0.090 | 0.3853 | No |
| 108 | DLG1 | 10400 | 0.090 | 0.3861 | No |
| 109 | SHROOM1 | 10478 | 0.089 | 0.3830 | No |
| 110 | KIFAP3 | 10526 | 0.088 | 0.3817 | No |
| 111 | EPB41 | 10538 | 0.088 | 0.3823 | No |
| 112 | ROCK1 | 10629 | 0.086 | 0.3785 | No |
| 113 | FLNB | 10799 | 0.083 | 0.3702 | No |
| 114 | DOCK2 | 10916 | 0.081 | 0.3648 | No |
| 115 | NEDD9 | 11074 | 0.078 | 0.3571 | No |
| 116 | MYH9 | 11259 | 0.075 | 0.3479 | No |
| 117 | PXN | 11294 | 0.075 | 0.3471 | No |
| 118 | PLK1 | 11329 | 0.074 | 0.3462 | No |
| 119 | PCGF5 | 11389 | 0.073 | 0.3439 | No |
| 120 | CLASP1 | 11442 | 0.073 | 0.3421 | No |
| 121 | CENPJ | 11518 | 0.071 | 0.3389 | No |
| 122 | CDC42EP1 | 11529 | 0.071 | 0.3393 | No |
| 123 | ARHGEF11 | 11570 | 0.071 | 0.3381 | No |
| 124 | KIF3B | 11786 | 0.067 | 0.3270 | No |
| 125 | ARHGEF7 | 11984 | 0.064 | 0.3169 | No |
| 126 | LMNB1 | 11987 | 0.064 | 0.3177 | No |
| 127 | NIN | 12000 | 0.064 | 0.3179 | No |
| 128 | NOTCH2 | 12003 | 0.064 | 0.3187 | No |
| 129 | CYTH2 | 12444 | 0.058 | 0.2948 | No |
| 130 | STK38L | 12523 | 0.057 | 0.2913 | No |
| 131 | RICTOR | 12526 | 0.057 | 0.2920 | No |
| 132 | LATS1 | 12716 | 0.054 | 0.2821 | No |
| 133 | RASA1 | 12842 | 0.052 | 0.2759 | No |
| 134 | KLC1 | 12914 | 0.051 | 0.2726 | No |
| 135 | CKAP5 | 12941 | 0.051 | 0.2719 | No |
| 136 | CENPF | 13109 | 0.049 | 0.2632 | No |
| 137 | ARHGEF3 | 13194 | 0.048 | 0.2592 | No |
| 138 | MID1 | 13255 | 0.047 | 0.2565 | No |
| 139 | CCDC88A | 13296 | 0.047 | 0.2549 | No |
| 140 | ARFIP2 | 13423 | 0.045 | 0.2485 | No |
| 141 | WASL | 13457 | 0.044 | 0.2473 | No |
| 142 | PREX1 | 13674 | 0.041 | 0.2357 | No |
| 143 | BCL2L11 | 13825 | 0.039 | 0.2279 | No |
| 144 | NCK2 | 13849 | 0.039 | 0.2272 | No |
| 145 | PCNT | 13919 | 0.038 | 0.2238 | No |
| 146 | TPX2 | 13929 | 0.037 | 0.2238 | No |
| 147 | SYNPO | 13986 | 0.036 | 0.2212 | No |
| 148 | KIF23 | 14136 | 0.033 | 0.2133 | No |
| 149 | MARK4 | 14230 | 0.031 | 0.2086 | No |
| 150 | RAB3GAP1 | 14272 | 0.030 | 0.2067 | No |
| 151 | CAPZB | 14429 | 0.021 | 0.1982 | No |
| 152 | YWHAE | 14513 | 0.014 | 0.1938 | No |
| 153 | CDC27 | 14542 | -0.000 | 0.1922 | No |
| 154 | PAFAH1B1 | 14561 | -0.000 | 0.1912 | No |
| 155 | RANBP9 | 14578 | -0.000 | 0.1903 | No |
| 156 | RABGAP1 | 14588 | -0.000 | 0.1898 | No |
| 157 | VCL | 14625 | -0.000 | 0.1878 | No |
| 158 | SMC1A | 14739 | -0.000 | 0.1814 | No |
| 159 | FSCN1 | 14757 | -0.000 | 0.1805 | No |
| 160 | OPHN1 | 14781 | -0.000 | 0.1792 | No |
| 161 | KIF3C | 14807 | -0.000 | 0.1778 | No |
| 162 | KIF4A | 14856 | -0.000 | 0.1751 | No |
| 163 | HDAC6 | 14870 | -0.000 | 0.1744 | No |
| 164 | MAPRE1 | 14954 | -0.000 | 0.1697 | No |
| 165 | CEP192 | 14963 | -0.000 | 0.1692 | No |
| 166 | WASF1 | 15241 | -0.000 | 0.1537 | No |
| 167 | SPTBN1 | 15315 | -0.000 | 0.1496 | No |
| 168 | STAU1 | 15534 | -0.000 | 0.1374 | No |
| 169 | UXT | 15606 | -0.000 | 0.1334 | No |
| 170 | PALLD | 15647 | -0.000 | 0.1311 | No |
| 171 | ABI1 | 15858 | -0.000 | 0.1193 | No |
| 172 | LRPPRC | 15904 | -0.000 | 0.1168 | No |
| 173 | KIF11 | 15907 | -0.000 | 0.1167 | No |
| 174 | ARHGDIA | 15991 | -0.000 | 0.1120 | No |
| 175 | CDC42BPA | 16042 | -0.000 | 0.1092 | No |
| 176 | ARL8A | 16044 | -0.000 | 0.1092 | No |
| 177 | NCK1 | 16316 | -0.000 | 0.0940 | No |
| 178 | WASF2 | 16319 | -0.000 | 0.0939 | No |
| 179 | PPP4R2 | 16467 | -0.000 | 0.0856 | No |
| 180 | MID1IP1 | 16552 | -0.000 | 0.0809 | No |
| 181 | TSC1 | 16576 | -0.000 | 0.0796 | No |
| 182 | CDK1 | 16779 | -0.000 | 0.0683 | No |
| 183 | CDC42EP4 | 17082 | -0.000 | 0.0513 | No |
| 184 | SEPTIN9 | 17244 | -0.000 | 0.0423 | No |
| 185 | FLNA | 17474 | -0.000 | 0.0294 | No |
| 186 | DYNC1H1 | 17483 | -0.000 | 0.0290 | No |
| 187 | TLK1 | 17552 | -0.000 | 0.0251 | No |
| 188 | PRC1 | 17577 | -0.000 | 0.0238 | No |
| 189 | DYNLL2 | 17895 | -0.000 | 0.0060 | No |
| 190 | MARCKS | 17959 | -0.000 | 0.0025 | No |