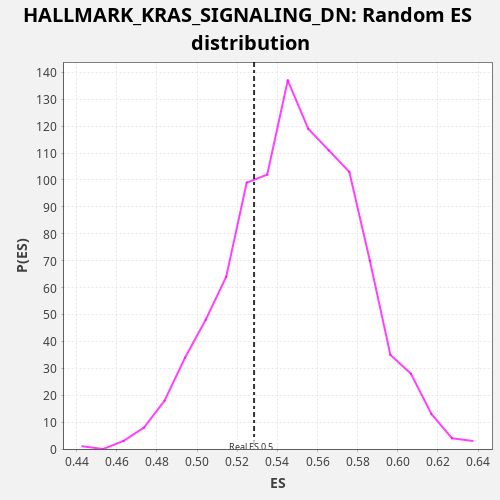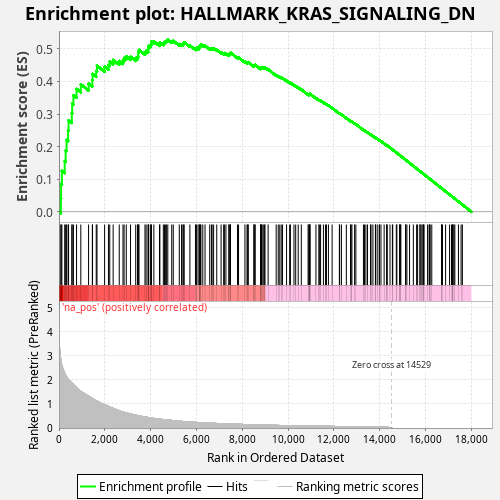
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.52838326 |
| Normalized Enrichment Score (NES) | 0.9625838 |
| Nominal p-value | 0.743 |
| FDR q-value | 0.88657445 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | ZNF112 | 81 | 2.850 | 0.0412 | Yes |
| 2 | MYH7 | 91 | 2.795 | 0.0856 | Yes |
| 3 | PRKN | 126 | 2.613 | 0.1257 | Yes |
| 4 | CELSR2 | 249 | 2.286 | 0.1556 | Yes |
| 5 | COQ8A | 291 | 2.193 | 0.1885 | Yes |
| 6 | CACNA1F | 331 | 2.135 | 0.2206 | Yes |
| 7 | LFNG | 397 | 2.032 | 0.2496 | Yes |
| 8 | SLC16A7 | 417 | 2.010 | 0.2808 | Yes |
| 9 | DCC | 559 | 1.871 | 0.3030 | Yes |
| 10 | CHST2 | 575 | 1.853 | 0.3319 | Yes |
| 11 | SLC38A3 | 630 | 1.800 | 0.3578 | Yes |
| 12 | KCNQ2 | 766 | 1.684 | 0.3773 | Yes |
| 13 | CDH16 | 953 | 1.528 | 0.3914 | Yes |
| 14 | SLC12A3 | 1291 | 1.325 | 0.3937 | Yes |
| 15 | GPR19 | 1455 | 1.225 | 0.4043 | Yes |
| 16 | KLHDC8A | 1464 | 1.220 | 0.4234 | Yes |
| 17 | ARPP21 | 1625 | 1.127 | 0.4325 | Yes |
| 18 | WNT16 | 1659 | 1.115 | 0.4486 | Yes |
| 19 | SCN10A | 1991 | 0.966 | 0.4455 | Yes |
| 20 | RGS11 | 2159 | 0.897 | 0.4506 | Yes |
| 21 | CYP39A1 | 2215 | 0.874 | 0.4615 | Yes |
| 22 | COPZ2 | 2364 | 0.815 | 0.4663 | Yes |
| 23 | EPHA5 | 2630 | 0.716 | 0.4629 | Yes |
| 24 | SELENOP | 2793 | 0.663 | 0.4645 | Yes |
| 25 | FGFR3 | 2841 | 0.651 | 0.4723 | Yes |
| 26 | ZC2HC1C | 2937 | 0.627 | 0.4770 | Yes |
| 27 | MYO15A | 3123 | 0.582 | 0.4760 | Yes |
| 28 | AKR1B10 | 3349 | 0.534 | 0.4719 | Yes |
| 29 | NPHS1 | 3426 | 0.518 | 0.4760 | Yes |
| 30 | CKM | 3458 | 0.512 | 0.4825 | Yes |
| 31 | EGF | 3459 | 0.512 | 0.4907 | Yes |
| 32 | CPB1 | 3493 | 0.504 | 0.4970 | Yes |
| 33 | IFI44L | 3753 | 0.457 | 0.4898 | Yes |
| 34 | PROP1 | 3803 | 0.449 | 0.4942 | Yes |
| 35 | ITIH3 | 3891 | 0.434 | 0.4963 | Yes |
| 36 | GAMT | 3896 | 0.434 | 0.5031 | Yes |
| 37 | PAX4 | 3914 | 0.431 | 0.5090 | Yes |
| 38 | SIDT1 | 3990 | 0.421 | 0.5116 | Yes |
| 39 | THNSL2 | 4026 | 0.417 | 0.5163 | Yes |
| 40 | ALOX12B | 4033 | 0.416 | 0.5227 | Yes |
| 41 | GTF3C5 | 4141 | 0.398 | 0.5231 | Yes |
| 42 | FGGY | 4388 | 0.368 | 0.5152 | Yes |
| 43 | PNMT | 4403 | 0.367 | 0.5203 | Yes |
| 44 | IDUA | 4555 | 0.349 | 0.5174 | Yes |
| 45 | CYP11B2 | 4604 | 0.345 | 0.5203 | Yes |
| 46 | ATP4A | 4644 | 0.341 | 0.5236 | Yes |
| 47 | CD207 | 4698 | 0.334 | 0.5260 | Yes |
| 48 | C5 | 4750 | 0.329 | 0.5284 | Yes |
| 49 | KRT15 | 4922 | 0.313 | 0.5238 | No |
| 50 | EDAR | 4988 | 0.306 | 0.5251 | No |
| 51 | UPK3B | 5251 | 0.283 | 0.5149 | No |
| 52 | TSHB | 5349 | 0.275 | 0.5139 | No |
| 53 | CPA2 | 5413 | 0.270 | 0.5147 | No |
| 54 | CAPN9 | 5424 | 0.269 | 0.5184 | No |
| 55 | SPTBN2 | 5466 | 0.266 | 0.5204 | No |
| 56 | TG | 5714 | 0.248 | 0.5105 | No |
| 57 | ENTPD7 | 5958 | 0.233 | 0.5006 | No |
| 58 | AMBN | 6015 | 0.230 | 0.5012 | No |
| 59 | KRT4 | 6016 | 0.230 | 0.5049 | No |
| 60 | TGM1 | 6115 | 0.225 | 0.5030 | No |
| 61 | ABCB11 | 6116 | 0.225 | 0.5066 | No |
| 62 | SLC29A3 | 6146 | 0.224 | 0.5086 | No |
| 63 | SERPINA10 | 6175 | 0.222 | 0.5106 | No |
| 64 | ATP6V1B1 | 6195 | 0.221 | 0.5131 | No |
| 65 | MYOT | 6277 | 0.217 | 0.5120 | No |
| 66 | RSAD2 | 6375 | 0.212 | 0.5100 | No |
| 67 | MTHFR | 6580 | 0.201 | 0.5017 | No |
| 68 | SLC5A5 | 6655 | 0.198 | 0.5008 | No |
| 69 | NRIP2 | 6684 | 0.196 | 0.5023 | No |
| 70 | SLC6A3 | 6750 | 0.193 | 0.5018 | No |
| 71 | VPS50 | 6887 | 0.187 | 0.4972 | No |
| 72 | TAS2R4 | 7076 | 0.179 | 0.4895 | No |
| 73 | KCNE2 | 7189 | 0.175 | 0.4860 | No |
| 74 | MAGIX | 7219 | 0.174 | 0.4872 | No |
| 75 | HNF1A | 7287 | 0.171 | 0.4862 | No |
| 76 | TEX15 | 7419 | 0.166 | 0.4815 | No |
| 77 | GPRC5C | 7420 | 0.166 | 0.4842 | No |
| 78 | SERPINB2 | 7465 | 0.165 | 0.4843 | No |
| 79 | MEFV | 7478 | 0.165 | 0.4863 | No |
| 80 | VPREB1 | 7485 | 0.165 | 0.4886 | No |
| 81 | MSH5 | 7793 | 0.154 | 0.4739 | No |
| 82 | ABCG4 | 7838 | 0.153 | 0.4739 | No |
| 83 | RIBC2 | 8117 | 0.143 | 0.4605 | No |
| 84 | SLC25A23 | 8207 | 0.141 | 0.4578 | No |
| 85 | CHRNG | 8237 | 0.139 | 0.4584 | No |
| 86 | MX1 | 8264 | 0.138 | 0.4592 | No |
| 87 | IL12B | 8510 | 0.132 | 0.4475 | No |
| 88 | PDK2 | 8512 | 0.132 | 0.4496 | No |
| 89 | NR6A1 | 8549 | 0.131 | 0.4497 | No |
| 90 | BARD1 | 8562 | 0.130 | 0.4511 | No |
| 91 | STAG3 | 8800 | 0.124 | 0.4398 | No |
| 92 | ITGB1BP2 | 8801 | 0.124 | 0.4418 | No |
| 93 | IFNG | 8819 | 0.124 | 0.4428 | No |
| 94 | TNNI3 | 8842 | 0.123 | 0.4436 | No |
| 95 | KRT5 | 8871 | 0.123 | 0.4440 | No |
| 96 | CDKAL1 | 8941 | 0.120 | 0.4420 | No |
| 97 | NOS1 | 8963 | 0.120 | 0.4428 | No |
| 98 | GP1BA | 8993 | 0.119 | 0.4431 | No |
| 99 | YPEL1 | 9129 | 0.116 | 0.4373 | No |
| 100 | IGFBP2 | 9481 | 0.108 | 0.4194 | No |
| 101 | KRT13 | 9571 | 0.106 | 0.4161 | No |
| 102 | PAX3 | 9638 | 0.104 | 0.4140 | No |
| 103 | UGT2B17 | 9701 | 0.103 | 0.4122 | No |
| 104 | KCNMB1 | 9759 | 0.102 | 0.4106 | No |
| 105 | ACTC1 | 9934 | 0.098 | 0.4024 | No |
| 106 | SPHK2 | 10069 | 0.096 | 0.3965 | No |
| 107 | SLC30A3 | 10102 | 0.095 | 0.3962 | No |
| 108 | CCR8 | 10258 | 0.092 | 0.3890 | No |
| 109 | MFSD6 | 10333 | 0.091 | 0.3863 | No |
| 110 | LYPD3 | 10443 | 0.089 | 0.3816 | No |
| 111 | BTG2 | 10578 | 0.087 | 0.3755 | No |
| 112 | HTR1D | 10874 | 0.081 | 0.3602 | No |
| 113 | EDN1 | 10914 | 0.081 | 0.3593 | No |
| 114 | CLSTN3 | 10918 | 0.081 | 0.3604 | No |
| 115 | NGB | 10937 | 0.080 | 0.3607 | No |
| 116 | P2RX6 | 10944 | 0.080 | 0.3617 | No |
| 117 | CALCB | 10962 | 0.080 | 0.3620 | No |
| 118 | CCDC106 | 11217 | 0.076 | 0.3490 | No |
| 119 | HSD11B2 | 11343 | 0.074 | 0.3431 | No |
| 120 | MAST3 | 11381 | 0.073 | 0.3422 | No |
| 121 | PDE6B | 11420 | 0.073 | 0.3413 | No |
| 122 | KRT1 | 11543 | 0.071 | 0.3356 | No |
| 123 | SGK1 | 11633 | 0.069 | 0.3317 | No |
| 124 | DLK2 | 11676 | 0.069 | 0.3304 | No |
| 125 | PCDHB1 | 11762 | 0.068 | 0.3267 | No |
| 126 | SSTR4 | 11923 | 0.065 | 0.3188 | No |
| 127 | KMT2D | 12240 | 0.061 | 0.3020 | No |
| 128 | FGF16 | 12251 | 0.061 | 0.3024 | No |
| 129 | DTNB | 12329 | 0.060 | 0.2991 | No |
| 130 | IL5 | 12544 | 0.056 | 0.2880 | No |
| 131 | CACNG1 | 12726 | 0.054 | 0.2787 | No |
| 132 | GPR3 | 12752 | 0.054 | 0.2781 | No |
| 133 | SLC6A14 | 12788 | 0.053 | 0.2770 | No |
| 134 | PTGFR | 12905 | 0.052 | 0.2713 | No |
| 135 | OXT | 12960 | 0.051 | 0.2691 | No |
| 136 | FSHB | 13291 | 0.047 | 0.2513 | No |
| 137 | KCND1 | 13339 | 0.046 | 0.2494 | No |
| 138 | CLDN16 | 13399 | 0.045 | 0.2469 | No |
| 139 | CNTFR | 13469 | 0.044 | 0.2437 | No |
| 140 | PDCD1 | 13598 | 0.042 | 0.2372 | No |
| 141 | SKIL | 13622 | 0.042 | 0.2366 | No |
| 142 | CD40LG | 13704 | 0.041 | 0.2327 | No |
| 143 | KLK7 | 13813 | 0.039 | 0.2272 | No |
| 144 | TCF7L1 | 13819 | 0.039 | 0.2276 | No |
| 145 | TFF2 | 13891 | 0.038 | 0.2242 | No |
| 146 | ARHGDIG | 13982 | 0.036 | 0.2197 | No |
| 147 | SYNPO | 13986 | 0.036 | 0.2202 | No |
| 148 | EFHD1 | 14056 | 0.035 | 0.2168 | No |
| 149 | KCNN1 | 14194 | 0.032 | 0.2097 | No |
| 150 | P2RY4 | 14307 | 0.028 | 0.2038 | No |
| 151 | ADRA2C | 14314 | 0.028 | 0.2040 | No |
| 152 | SMPX | 14324 | 0.028 | 0.2039 | No |
| 153 | NR4A2 | 14450 | 0.020 | 0.1972 | No |
| 154 | YBX2 | 14554 | -0.000 | 0.1914 | No |
| 155 | TFAP2B | 14569 | -0.000 | 0.1906 | No |
| 156 | FGF22 | 14726 | -0.000 | 0.1819 | No |
| 157 | SNCB | 14750 | -0.000 | 0.1806 | No |
| 158 | TGFB2 | 14869 | -0.000 | 0.1739 | No |
| 159 | MACROH2A2 | 14884 | -0.000 | 0.1732 | No |
| 160 | SOX10 | 14899 | -0.000 | 0.1724 | No |
| 161 | TCL1A | 14924 | -0.000 | 0.1710 | No |
| 162 | TLX1 | 15134 | -0.000 | 0.1593 | No |
| 163 | CPEB3 | 15136 | -0.000 | 0.1592 | No |
| 164 | ZBTB16 | 15188 | -0.000 | 0.1564 | No |
| 165 | TFCP2L1 | 15304 | -0.000 | 0.1499 | No |
| 166 | CD80 | 15469 | -0.000 | 0.1407 | No |
| 167 | EDN2 | 15612 | -0.000 | 0.1327 | No |
| 168 | KLK8 | 15655 | -0.000 | 0.1304 | No |
| 169 | CCNA1 | 15758 | -0.000 | 0.1246 | No |
| 170 | IRS4 | 15762 | -0.000 | 0.1245 | No |
| 171 | HTR1B | 15812 | -0.000 | 0.1217 | No |
| 172 | CLPS | 15887 | -0.000 | 0.1176 | No |
| 173 | BRDT | 15897 | -0.000 | 0.1171 | No |
| 174 | BMPR1B | 15919 | -0.000 | 0.1159 | No |
| 175 | COL2A1 | 15935 | -0.000 | 0.1150 | No |
| 176 | TENM2 | 16091 | -0.000 | 0.1063 | No |
| 177 | PTPRJ | 16157 | -0.000 | 0.1027 | No |
| 178 | THRB | 16181 | -0.000 | 0.1014 | No |
| 179 | GRID2 | 16203 | -0.000 | 0.1002 | No |
| 180 | CLDN8 | 16274 | -0.000 | 0.0963 | No |
| 181 | GDNF | 16702 | -0.000 | 0.0723 | No |
| 182 | SHOX2 | 16710 | -0.000 | 0.0719 | No |
| 183 | GP2 | 16741 | -0.000 | 0.0702 | No |
| 184 | INSL5 | 16877 | -0.000 | 0.0627 | No |
| 185 | CALML5 | 17051 | -0.000 | 0.0529 | No |
| 186 | PLAG1 | 17134 | -0.000 | 0.0483 | No |
| 187 | CAMK1D | 17180 | -0.000 | 0.0458 | No |
| 188 | ASB7 | 17193 | -0.000 | 0.0451 | No |
| 189 | TENT5C | 17196 | -0.000 | 0.0450 | No |
| 190 | SNN | 17243 | -0.000 | 0.0424 | No |
| 191 | NTF3 | 17281 | -0.000 | 0.0404 | No |
| 192 | NUDT11 | 17438 | -0.000 | 0.0316 | No |
| 193 | RYR2 | 17554 | -0.000 | 0.0252 | No |
| 194 | NPY4R | 17610 | -0.000 | 0.0221 | No |