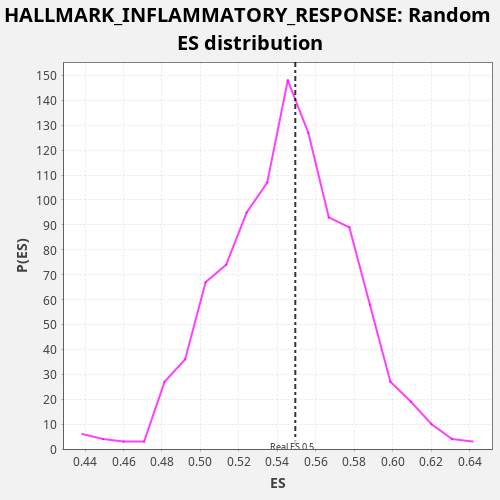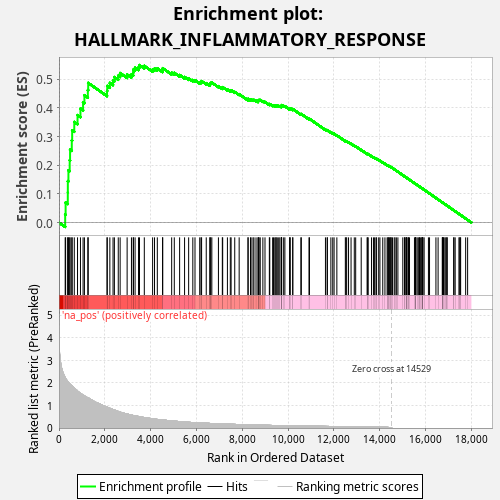
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | 0.5493392 |
| Normalized Enrichment Score (NES) | 1.008654 |
| Nominal p-value | 0.448 |
| FDR q-value | 0.80248266 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | NFKBIA | 270 | 2.233 | 0.0283 | Yes |
| 2 | SCN1B | 289 | 2.196 | 0.0701 | Yes |
| 3 | MMP14 | 384 | 2.049 | 0.1047 | Yes |
| 4 | PSEN1 | 386 | 2.048 | 0.1445 | Yes |
| 5 | CXCR6 | 412 | 2.018 | 0.1824 | Yes |
| 6 | F3 | 467 | 1.956 | 0.2174 | Yes |
| 7 | ACVR2A | 484 | 1.935 | 0.2542 | Yes |
| 8 | GABBR1 | 558 | 1.871 | 0.2865 | Yes |
| 9 | CHST2 | 575 | 1.853 | 0.3217 | Yes |
| 10 | KCNA3 | 667 | 1.762 | 0.3509 | Yes |
| 11 | MET | 809 | 1.633 | 0.3748 | Yes |
| 12 | SLC31A2 | 937 | 1.539 | 0.3976 | Yes |
| 13 | SGMS2 | 1051 | 1.464 | 0.4198 | Yes |
| 14 | IL18RAP | 1110 | 1.421 | 0.4442 | Yes |
| 15 | CLEC5A | 1259 | 1.343 | 0.4620 | Yes |
| 16 | SLC4A4 | 1275 | 1.333 | 0.4871 | Yes |
| 17 | LDLR | 2097 | 0.924 | 0.4590 | Yes |
| 18 | IL1A | 2106 | 0.921 | 0.4765 | Yes |
| 19 | PIK3R5 | 2220 | 0.873 | 0.4872 | Yes |
| 20 | ADORA2B | 2359 | 0.818 | 0.4953 | Yes |
| 21 | IL18R1 | 2418 | 0.795 | 0.5076 | Yes |
| 22 | IRF7 | 2587 | 0.732 | 0.5124 | Yes |
| 23 | BEST1 | 2669 | 0.701 | 0.5215 | Yes |
| 24 | TPBG | 2974 | 0.618 | 0.5164 | Yes |
| 25 | ITGB3 | 3163 | 0.573 | 0.5170 | Yes |
| 26 | CSF3R | 3242 | 0.554 | 0.5235 | Yes |
| 27 | CXCL9 | 3248 | 0.553 | 0.5339 | Yes |
| 28 | NPFFR2 | 3322 | 0.539 | 0.5403 | Yes |
| 29 | CD82 | 3472 | 0.509 | 0.5419 | Yes |
| 30 | SLC28A2 | 3514 | 0.501 | 0.5493 | Yes |
| 31 | LAMP3 | 3724 | 0.461 | 0.5466 | No |
| 32 | TACR1 | 4087 | 0.407 | 0.5342 | No |
| 33 | IFNAR1 | 4170 | 0.394 | 0.5372 | No |
| 34 | TLR1 | 4292 | 0.379 | 0.5378 | No |
| 35 | DCBLD2 | 4521 | 0.353 | 0.5319 | No |
| 36 | SCARF1 | 4528 | 0.352 | 0.5384 | No |
| 37 | ABCA1 | 4919 | 0.313 | 0.5226 | No |
| 38 | P2RX7 | 5033 | 0.303 | 0.5222 | No |
| 39 | SELE | 5266 | 0.282 | 0.5146 | No |
| 40 | CXCL8 | 5482 | 0.265 | 0.5077 | No |
| 41 | CCL24 | 5660 | 0.252 | 0.5027 | No |
| 42 | SELL | 5842 | 0.240 | 0.4972 | No |
| 43 | TNFSF10 | 5937 | 0.234 | 0.4965 | No |
| 44 | ADGRE1 | 6143 | 0.224 | 0.4893 | No |
| 45 | CCL5 | 6199 | 0.221 | 0.4906 | No |
| 46 | MARCO | 6225 | 0.220 | 0.4934 | No |
| 47 | PVR | 6428 | 0.209 | 0.4862 | No |
| 48 | STAB1 | 6585 | 0.201 | 0.4813 | No |
| 49 | TLR3 | 6596 | 0.201 | 0.4847 | No |
| 50 | CD48 | 6624 | 0.199 | 0.4870 | No |
| 51 | CD55 | 6672 | 0.197 | 0.4882 | No |
| 52 | ICAM4 | 6960 | 0.185 | 0.4757 | No |
| 53 | P2RX4 | 7130 | 0.177 | 0.4697 | No |
| 54 | LCK | 7146 | 0.176 | 0.4722 | No |
| 55 | TACR3 | 7352 | 0.169 | 0.4640 | No |
| 56 | MEFV | 7478 | 0.165 | 0.4602 | No |
| 57 | CXCL6 | 7513 | 0.163 | 0.4615 | No |
| 58 | CALCRL | 7672 | 0.158 | 0.4557 | No |
| 59 | LCP2 | 7865 | 0.152 | 0.4479 | No |
| 60 | EIF2AK2 | 8251 | 0.139 | 0.4290 | No |
| 61 | ICAM1 | 8256 | 0.139 | 0.4315 | No |
| 62 | NMUR1 | 8360 | 0.136 | 0.4283 | No |
| 63 | NOD2 | 8386 | 0.135 | 0.4295 | No |
| 64 | TNFRSF9 | 8469 | 0.133 | 0.4275 | No |
| 65 | IL12B | 8510 | 0.132 | 0.4278 | No |
| 66 | SLC31A1 | 8601 | 0.129 | 0.4253 | No |
| 67 | AXL | 8690 | 0.127 | 0.4228 | No |
| 68 | FZD5 | 8699 | 0.127 | 0.4248 | No |
| 69 | EREG | 8706 | 0.127 | 0.4270 | No |
| 70 | SLC7A2 | 8738 | 0.126 | 0.4277 | No |
| 71 | KIF1B | 8785 | 0.125 | 0.4275 | No |
| 72 | IL7R | 8900 | 0.122 | 0.4235 | No |
| 73 | GP1BA | 8993 | 0.119 | 0.4207 | No |
| 74 | CDKN1A | 9183 | 0.114 | 0.4123 | No |
| 75 | APLNR | 9201 | 0.114 | 0.4135 | No |
| 76 | NLRP3 | 9327 | 0.111 | 0.4087 | No |
| 77 | OLR1 | 9363 | 0.110 | 0.4089 | No |
| 78 | EBI3 | 9421 | 0.109 | 0.4078 | No |
| 79 | RGS1 | 9468 | 0.108 | 0.4073 | No |
| 80 | RNF144B | 9479 | 0.108 | 0.4088 | No |
| 81 | IL15RA | 9529 | 0.107 | 0.4081 | No |
| 82 | TAPBP | 9592 | 0.105 | 0.4067 | No |
| 83 | CCL22 | 9641 | 0.104 | 0.4061 | No |
| 84 | IFNGR2 | 9703 | 0.103 | 0.4046 | No |
| 85 | AQP9 | 9712 | 0.103 | 0.4062 | No |
| 86 | SRI | 9713 | 0.103 | 0.4082 | No |
| 87 | AHR | 9723 | 0.102 | 0.4097 | No |
| 88 | SPHK1 | 9808 | 0.100 | 0.4069 | No |
| 89 | HIF1A | 9861 | 0.099 | 0.4059 | No |
| 90 | GNAI3 | 10073 | 0.096 | 0.3959 | No |
| 91 | ITGA5 | 10076 | 0.096 | 0.3977 | No |
| 92 | PDPN | 10090 | 0.095 | 0.3988 | No |
| 93 | PTGER2 | 10204 | 0.093 | 0.3943 | No |
| 94 | EMP3 | 10206 | 0.093 | 0.3960 | No |
| 95 | RASGRP1 | 10560 | 0.087 | 0.3779 | No |
| 96 | BTG2 | 10578 | 0.087 | 0.3787 | No |
| 97 | EDN1 | 10914 | 0.081 | 0.3614 | No |
| 98 | CCL17 | 10935 | 0.080 | 0.3619 | No |
| 99 | CD69 | 11632 | 0.069 | 0.3242 | No |
| 100 | ITGB8 | 11708 | 0.068 | 0.3213 | No |
| 101 | PTGIR | 11715 | 0.068 | 0.3223 | No |
| 102 | BST2 | 11869 | 0.066 | 0.3150 | No |
| 103 | SELENOS | 11951 | 0.064 | 0.3117 | No |
| 104 | PLAUR | 12009 | 0.064 | 0.3097 | No |
| 105 | PTAFR | 12130 | 0.062 | 0.3042 | No |
| 106 | ATP2C1 | 12500 | 0.057 | 0.2846 | No |
| 107 | OSMR | 12528 | 0.057 | 0.2842 | No |
| 108 | RIPK2 | 12559 | 0.056 | 0.2836 | No |
| 109 | TNFRSF1B | 12640 | 0.055 | 0.2802 | No |
| 110 | IL10RA | 12750 | 0.054 | 0.2751 | No |
| 111 | GNA15 | 12891 | 0.052 | 0.2682 | No |
| 112 | CD40 | 12952 | 0.051 | 0.2659 | No |
| 113 | IL1R1 | 13193 | 0.048 | 0.2533 | No |
| 114 | IL2RB | 13455 | 0.044 | 0.2395 | No |
| 115 | ROS1 | 13461 | 0.044 | 0.2401 | No |
| 116 | CCR7 | 13499 | 0.044 | 0.2389 | No |
| 117 | ACVR1B | 13644 | 0.042 | 0.2316 | No |
| 118 | IL4R | 13728 | 0.041 | 0.2277 | No |
| 119 | IL15 | 13778 | 0.040 | 0.2258 | No |
| 120 | RGS16 | 13838 | 0.039 | 0.2232 | No |
| 121 | SLC11A2 | 13865 | 0.039 | 0.2225 | No |
| 122 | TNFSF15 | 13957 | 0.037 | 0.2181 | No |
| 123 | PTGER4 | 14010 | 0.036 | 0.2159 | No |
| 124 | SLAMF1 | 14134 | 0.034 | 0.2096 | No |
| 125 | SEMA4D | 14229 | 0.031 | 0.2050 | No |
| 126 | P2RY2 | 14325 | 0.028 | 0.2002 | No |
| 127 | INHBA | 14354 | 0.026 | 0.1991 | No |
| 128 | GPR132 | 14395 | 0.022 | 0.1973 | No |
| 129 | OSM | 14398 | 0.022 | 0.1976 | No |
| 130 | KLF6 | 14423 | 0.021 | 0.1967 | No |
| 131 | RHOG | 14483 | 0.017 | 0.1937 | No |
| 132 | ADM | 14493 | 0.016 | 0.1935 | No |
| 133 | FFAR2 | 14496 | 0.016 | 0.1937 | No |
| 134 | CX3CL1 | 14558 | -0.000 | 0.1903 | No |
| 135 | MSR1 | 14633 | -0.000 | 0.1861 | No |
| 136 | MXD1 | 14670 | -0.000 | 0.1841 | No |
| 137 | ATP2B1 | 14728 | -0.000 | 0.1809 | No |
| 138 | OPRK1 | 14795 | -0.000 | 0.1772 | No |
| 139 | TIMP1 | 15009 | -0.000 | 0.1653 | No |
| 140 | HPN | 15082 | -0.000 | 0.1612 | No |
| 141 | NAMPT | 15086 | -0.000 | 0.1610 | No |
| 142 | CSF3 | 15150 | -0.000 | 0.1575 | No |
| 143 | CCL7 | 15157 | -0.000 | 0.1572 | No |
| 144 | CCL2 | 15158 | -0.000 | 0.1572 | No |
| 145 | NFKB1 | 15179 | -0.000 | 0.1560 | No |
| 146 | SLC1A2 | 15196 | -0.000 | 0.1551 | No |
| 147 | MEP1A | 15256 | -0.000 | 0.1518 | No |
| 148 | HBEGF | 15262 | -0.000 | 0.1516 | No |
| 149 | CCL20 | 15302 | -0.000 | 0.1494 | No |
| 150 | KCNJ2 | 15518 | -0.000 | 0.1373 | No |
| 151 | IRF1 | 15558 | -0.000 | 0.1351 | No |
| 152 | IL1B | 15568 | -0.000 | 0.1346 | No |
| 153 | TNFSF9 | 15571 | -0.000 | 0.1345 | No |
| 154 | CD70 | 15573 | -0.000 | 0.1344 | No |
| 155 | LIF | 15635 | -0.000 | 0.1310 | No |
| 156 | ADRM1 | 15686 | -0.000 | 0.1282 | No |
| 157 | GCH1 | 15732 | -0.000 | 0.1257 | No |
| 158 | RAF1 | 15736 | -0.000 | 0.1255 | No |
| 159 | IRAK2 | 15776 | -0.000 | 0.1233 | No |
| 160 | IL6 | 15843 | -0.000 | 0.1196 | No |
| 161 | RTP4 | 15852 | -0.000 | 0.1192 | No |
| 162 | IL10 | 15855 | -0.000 | 0.1191 | No |
| 163 | ABI1 | 15858 | -0.000 | 0.1189 | No |
| 164 | MYC | 15873 | -0.000 | 0.1182 | No |
| 165 | TLR2 | 15888 | -0.000 | 0.1174 | No |
| 166 | SLC7A1 | 15940 | -0.000 | 0.1145 | No |
| 167 | GPC3 | 16128 | -0.000 | 0.1040 | No |
| 168 | IL18 | 16176 | -0.000 | 0.1014 | No |
| 169 | PROK2 | 16455 | -0.000 | 0.0858 | No |
| 170 | CYBB | 16550 | -0.000 | 0.0805 | No |
| 171 | CXCL10 | 16736 | -0.000 | 0.0701 | No |
| 172 | CXCL11 | 16737 | -0.000 | 0.0701 | No |
| 173 | GPR183 | 16748 | -0.000 | 0.0695 | No |
| 174 | PCDH7 | 16761 | -0.000 | 0.0689 | No |
| 175 | CD14 | 16786 | -0.000 | 0.0675 | No |
| 176 | HAS2 | 16809 | -0.000 | 0.0663 | No |
| 177 | C3AR1 | 16859 | -0.000 | 0.0635 | No |
| 178 | RELA | 16902 | -0.000 | 0.0612 | No |
| 179 | ATP2A2 | 16936 | -0.000 | 0.0593 | No |
| 180 | CMKLR1 | 16947 | -0.000 | 0.0588 | No |
| 181 | CSF1 | 17230 | -0.000 | 0.0429 | No |
| 182 | PDE4B | 17242 | -0.000 | 0.0423 | No |
| 183 | IFITM1 | 17295 | -0.000 | 0.0394 | No |
| 184 | HRH1 | 17460 | -0.000 | 0.0302 | No |
| 185 | C5AR1 | 17501 | -0.000 | 0.0280 | No |
| 186 | KCNMB2 | 17506 | -0.000 | 0.0277 | No |
| 187 | LPAR1 | 17534 | -0.000 | 0.0262 | No |
| 188 | LTA | 17753 | -0.000 | 0.0140 | No |
| 189 | LYN | 17839 | -0.000 | 0.0092 | No |