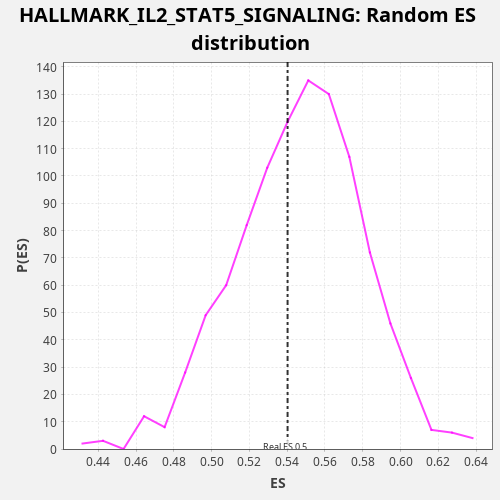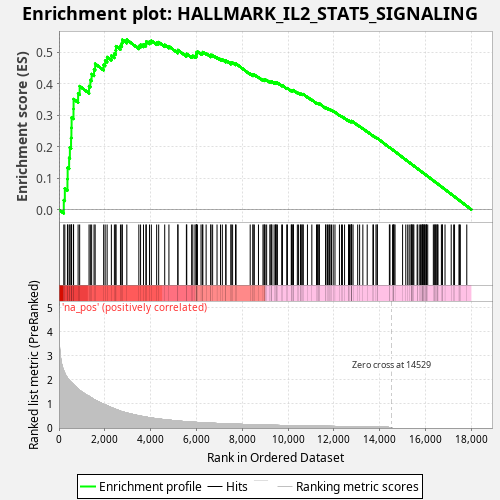
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_IL2_STAT5_SIGNALING |
| Enrichment Score (ES) | 0.540179 |
| Normalized Enrichment Score (NES) | 0.9883626 |
| Nominal p-value | 0.591 |
| FDR q-value | 0.8298854 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | CCNE1 | 209 | 2.370 | 0.0300 | Yes |
| 2 | CD81 | 255 | 2.255 | 0.0672 | Yes |
| 3 | BMPR2 | 369 | 2.067 | 0.0973 | Yes |
| 4 | MAFF | 378 | 2.060 | 0.1332 | Yes |
| 5 | CCND3 | 442 | 1.984 | 0.1646 | Yes |
| 6 | CCR4 | 475 | 1.949 | 0.1972 | Yes |
| 7 | WLS | 527 | 1.898 | 0.2278 | Yes |
| 8 | CAPN3 | 542 | 1.888 | 0.2602 | Yes |
| 9 | FURIN | 551 | 1.878 | 0.2929 | Yes |
| 10 | PLAGL1 | 633 | 1.795 | 0.3200 | Yes |
| 11 | CASP3 | 637 | 1.789 | 0.3513 | Yes |
| 12 | IL2RA | 834 | 1.615 | 0.3688 | Yes |
| 13 | PLSCR1 | 902 | 1.560 | 0.3925 | Yes |
| 14 | RABGAP1L | 1312 | 1.315 | 0.3927 | Yes |
| 15 | CDC6 | 1368 | 1.274 | 0.4121 | Yes |
| 16 | CYFIP1 | 1425 | 1.248 | 0.4309 | Yes |
| 17 | IGF1R | 1530 | 1.180 | 0.4459 | Yes |
| 18 | CDCP1 | 1578 | 1.159 | 0.4637 | Yes |
| 19 | RNH1 | 1950 | 0.982 | 0.4602 | Yes |
| 20 | SCN9A | 2014 | 0.958 | 0.4735 | Yes |
| 21 | ADAM19 | 2101 | 0.922 | 0.4850 | Yes |
| 22 | PTRH2 | 2286 | 0.844 | 0.4895 | Yes |
| 23 | IL18R1 | 2418 | 0.795 | 0.4962 | Yes |
| 24 | SPP1 | 2477 | 0.773 | 0.5065 | Yes |
| 25 | NCOA3 | 2488 | 0.769 | 0.5195 | Yes |
| 26 | MAP6 | 2683 | 0.699 | 0.5209 | Yes |
| 27 | SHE | 2744 | 0.677 | 0.5295 | Yes |
| 28 | ABCB1 | 2765 | 0.671 | 0.5402 | Yes |
| 29 | SNX14 | 2961 | 0.621 | 0.5402 | No |
| 30 | AGER | 3486 | 0.505 | 0.5197 | No |
| 31 | COCH | 3561 | 0.492 | 0.5242 | No |
| 32 | ECM1 | 3688 | 0.467 | 0.5253 | No |
| 33 | SMPDL3A | 3794 | 0.450 | 0.5274 | No |
| 34 | ENO3 | 3811 | 0.447 | 0.5344 | No |
| 35 | ETV4 | 3950 | 0.426 | 0.5341 | No |
| 36 | TTC39B | 4031 | 0.417 | 0.5370 | No |
| 37 | IL1R2 | 4263 | 0.382 | 0.5307 | No |
| 38 | DCPS | 4352 | 0.372 | 0.5323 | No |
| 39 | SELP | 4615 | 0.344 | 0.5237 | No |
| 40 | CAPG | 4799 | 0.325 | 0.5191 | No |
| 41 | SLC29A2 | 5187 | 0.289 | 0.5025 | No |
| 42 | ENPP1 | 5193 | 0.288 | 0.5073 | No |
| 43 | TNFRSF4 | 5561 | 0.260 | 0.4913 | No |
| 44 | PHTF2 | 5577 | 0.258 | 0.4950 | No |
| 45 | GADD45B | 5791 | 0.243 | 0.4873 | No |
| 46 | SELL | 5842 | 0.240 | 0.4887 | No |
| 47 | TNFSF10 | 5937 | 0.234 | 0.4876 | No |
| 48 | SLC2A3 | 5992 | 0.232 | 0.4886 | No |
| 49 | CST7 | 5993 | 0.232 | 0.4927 | No |
| 50 | EOMES | 6002 | 0.231 | 0.4963 | No |
| 51 | PLIN2 | 6004 | 0.231 | 0.5004 | No |
| 52 | FAH | 6044 | 0.229 | 0.5022 | No |
| 53 | ANXA4 | 6200 | 0.221 | 0.4974 | No |
| 54 | GBP4 | 6259 | 0.218 | 0.4980 | No |
| 55 | ITIH5 | 6280 | 0.217 | 0.5007 | No |
| 56 | TNFRSF18 | 6423 | 0.209 | 0.4964 | No |
| 57 | CD48 | 6624 | 0.199 | 0.4887 | No |
| 58 | PTCH1 | 6632 | 0.199 | 0.4918 | No |
| 59 | MUC1 | 6707 | 0.195 | 0.4911 | No |
| 60 | ITGA6 | 6899 | 0.187 | 0.4836 | No |
| 61 | LRIG1 | 7051 | 0.180 | 0.4783 | No |
| 62 | P2RX4 | 7130 | 0.177 | 0.4771 | No |
| 63 | MYO1E | 7281 | 0.171 | 0.4717 | No |
| 64 | PHLDA1 | 7288 | 0.171 | 0.4744 | No |
| 65 | NDRG1 | 7505 | 0.164 | 0.4651 | No |
| 66 | IL1RL1 | 7515 | 0.163 | 0.4675 | No |
| 67 | GPR83 | 7579 | 0.161 | 0.4668 | No |
| 68 | LRRC8C | 7706 | 0.157 | 0.4625 | No |
| 69 | PNP | 7730 | 0.156 | 0.4640 | No |
| 70 | CTSZ | 8347 | 0.136 | 0.4318 | No |
| 71 | IL3RA | 8468 | 0.133 | 0.4274 | No |
| 72 | TNFRSF9 | 8469 | 0.133 | 0.4297 | No |
| 73 | CISH | 8527 | 0.131 | 0.4288 | No |
| 74 | LCLAT1 | 8708 | 0.126 | 0.4210 | No |
| 75 | PLPP1 | 8905 | 0.121 | 0.4121 | No |
| 76 | SPRED2 | 8956 | 0.120 | 0.4114 | No |
| 77 | NT5E | 8962 | 0.120 | 0.4132 | No |
| 78 | APLP1 | 8998 | 0.119 | 0.4134 | No |
| 79 | HK2 | 9065 | 0.118 | 0.4117 | No |
| 80 | TIAM1 | 9204 | 0.114 | 0.4060 | No |
| 81 | GPX4 | 9255 | 0.112 | 0.4052 | No |
| 82 | RORA | 9263 | 0.112 | 0.4067 | No |
| 83 | EEF1AKMT1 | 9296 | 0.112 | 0.4069 | No |
| 84 | SNX9 | 9391 | 0.109 | 0.4036 | No |
| 85 | BATF | 9451 | 0.108 | 0.4021 | No |
| 86 | BATF3 | 9477 | 0.108 | 0.4026 | No |
| 87 | DENND5A | 9478 | 0.108 | 0.4045 | No |
| 88 | GUCY1B1 | 9535 | 0.107 | 0.4033 | No |
| 89 | AHR | 9723 | 0.102 | 0.3946 | No |
| 90 | AMACR | 9755 | 0.102 | 0.3946 | No |
| 91 | IFITM3 | 9943 | 0.098 | 0.3859 | No |
| 92 | AHCY | 9972 | 0.097 | 0.3860 | No |
| 93 | COL6A1 | 10139 | 0.094 | 0.3783 | No |
| 94 | ICOS | 10152 | 0.094 | 0.3793 | No |
| 95 | PTGER2 | 10204 | 0.093 | 0.3781 | No |
| 96 | SH3BGRL2 | 10219 | 0.093 | 0.3790 | No |
| 97 | PDCD2L | 10235 | 0.093 | 0.3797 | No |
| 98 | ODC1 | 10405 | 0.090 | 0.3718 | No |
| 99 | NFKBIZ | 10459 | 0.089 | 0.3704 | No |
| 100 | ITGAE | 10542 | 0.087 | 0.3674 | No |
| 101 | GSTO1 | 10562 | 0.087 | 0.3678 | No |
| 102 | GABARAPL1 | 10601 | 0.086 | 0.3672 | No |
| 103 | XBP1 | 10646 | 0.086 | 0.3663 | No |
| 104 | TWSG1 | 10661 | 0.085 | 0.3670 | No |
| 105 | PTH1R | 10846 | 0.082 | 0.3581 | No |
| 106 | POU2F1 | 11038 | 0.078 | 0.3488 | No |
| 107 | ITGAV | 11241 | 0.076 | 0.3387 | No |
| 108 | TRAF1 | 11280 | 0.075 | 0.3379 | No |
| 109 | CD83 | 11310 | 0.074 | 0.3376 | No |
| 110 | ST3GAL4 | 11350 | 0.074 | 0.3367 | No |
| 111 | PRKCH | 11367 | 0.074 | 0.3371 | No |
| 112 | UMPS | 11637 | 0.069 | 0.3232 | No |
| 113 | IFNGR1 | 11655 | 0.069 | 0.3235 | No |
| 114 | DRC1 | 11723 | 0.068 | 0.3209 | No |
| 115 | GLIPR2 | 11752 | 0.068 | 0.3206 | No |
| 116 | HOPX | 11814 | 0.067 | 0.3183 | No |
| 117 | NRP1 | 11833 | 0.066 | 0.3185 | No |
| 118 | PUS1 | 11897 | 0.065 | 0.3161 | No |
| 119 | SYNGR2 | 11970 | 0.064 | 0.3132 | No |
| 120 | IRF4 | 12048 | 0.063 | 0.3100 | No |
| 121 | SWAP70 | 12243 | 0.061 | 0.3001 | No |
| 122 | CA2 | 12332 | 0.060 | 0.2963 | No |
| 123 | ALCAM | 12373 | 0.059 | 0.2950 | No |
| 124 | TNFRSF8 | 12467 | 0.058 | 0.2908 | No |
| 125 | TNFRSF1B | 12640 | 0.055 | 0.2822 | No |
| 126 | IKZF4 | 12654 | 0.055 | 0.2824 | No |
| 127 | FLT3LG | 12682 | 0.055 | 0.2818 | No |
| 128 | IL10RA | 12750 | 0.054 | 0.2790 | No |
| 129 | TGM2 | 12754 | 0.054 | 0.2798 | No |
| 130 | P4HA1 | 12758 | 0.053 | 0.2806 | No |
| 131 | EMP1 | 12773 | 0.053 | 0.2807 | No |
| 132 | PRNP | 12776 | 0.053 | 0.2816 | No |
| 133 | LTB | 12838 | 0.052 | 0.2791 | No |
| 134 | SPRY4 | 13039 | 0.050 | 0.2687 | No |
| 135 | FGL2 | 13124 | 0.049 | 0.2648 | No |
| 136 | SLC1A5 | 13265 | 0.047 | 0.2578 | No |
| 137 | IL2RB | 13455 | 0.044 | 0.2480 | No |
| 138 | HIPK2 | 13705 | 0.041 | 0.2347 | No |
| 139 | IL4R | 13728 | 0.041 | 0.2342 | No |
| 140 | RGS16 | 13838 | 0.039 | 0.2288 | No |
| 141 | HUWE1 | 13892 | 0.038 | 0.2265 | No |
| 142 | SERPINB6 | 13894 | 0.038 | 0.2271 | No |
| 143 | KLF6 | 14423 | 0.021 | 0.1978 | No |
| 144 | BMP2 | 14448 | 0.020 | 0.1968 | No |
| 145 | CD79B | 14562 | -0.000 | 0.1905 | No |
| 146 | RRAGD | 14611 | -0.000 | 0.1878 | No |
| 147 | CD44 | 14614 | -0.000 | 0.1876 | No |
| 148 | IKZF2 | 14620 | -0.000 | 0.1874 | No |
| 149 | MXD1 | 14670 | -0.000 | 0.1846 | No |
| 150 | GATA1 | 15000 | -0.000 | 0.1661 | No |
| 151 | MAP3K8 | 15139 | -0.000 | 0.1584 | No |
| 152 | NOP2 | 15224 | -0.000 | 0.1537 | No |
| 153 | CD86 | 15288 | -0.000 | 0.1501 | No |
| 154 | IRF6 | 15372 | -0.000 | 0.1455 | No |
| 155 | SERPINC1 | 15374 | -0.000 | 0.1454 | No |
| 156 | CCND2 | 15397 | -0.000 | 0.1442 | No |
| 157 | TNFSF11 | 15445 | -0.000 | 0.1415 | No |
| 158 | SOCS2 | 15453 | -0.000 | 0.1412 | No |
| 159 | ARL4A | 15491 | -0.000 | 0.1391 | No |
| 160 | LIF | 15635 | -0.000 | 0.1311 | No |
| 161 | CDKN1C | 15664 | -0.000 | 0.1295 | No |
| 162 | SYT11 | 15751 | -0.000 | 0.1246 | No |
| 163 | BHLHE40 | 15778 | -0.000 | 0.1232 | No |
| 164 | CKAP4 | 15837 | -0.000 | 0.1199 | No |
| 165 | IL10 | 15855 | -0.000 | 0.1190 | No |
| 166 | MYC | 15873 | -0.000 | 0.1180 | No |
| 167 | PIM1 | 15881 | -0.000 | 0.1176 | No |
| 168 | SLC39A8 | 15928 | -0.000 | 0.1150 | No |
| 169 | ETFBKMT | 15932 | -0.000 | 0.1149 | No |
| 170 | GPR65 | 15962 | -0.000 | 0.1133 | No |
| 171 | IRF8 | 15981 | -0.000 | 0.1122 | No |
| 172 | UCK2 | 16021 | -0.000 | 0.1101 | No |
| 173 | RHOB | 16045 | -0.000 | 0.1088 | No |
| 174 | GALM | 16046 | -0.000 | 0.1088 | No |
| 175 | TNFRSF21 | 16097 | -0.000 | 0.1060 | No |
| 176 | CDC42SE2 | 16342 | -0.000 | 0.0923 | No |
| 177 | S100A1 | 16380 | -0.000 | 0.0902 | No |
| 178 | DHRS3 | 16413 | -0.000 | 0.0884 | No |
| 179 | MAPKAPK2 | 16428 | -0.000 | 0.0876 | No |
| 180 | CTLA4 | 16465 | -0.000 | 0.0856 | No |
| 181 | F2RL2 | 16508 | -0.000 | 0.0832 | No |
| 182 | CSF2 | 16515 | -0.000 | 0.0829 | No |
| 183 | NFIL3 | 16546 | -0.000 | 0.0812 | No |
| 184 | RHOH | 16694 | -0.000 | 0.0729 | No |
| 185 | IL13 | 16732 | -0.000 | 0.0709 | No |
| 186 | CXCL10 | 16736 | -0.000 | 0.0707 | No |
| 187 | BCL2 | 16852 | -0.000 | 0.0642 | No |
| 188 | PENK | 17122 | -0.000 | 0.0491 | No |
| 189 | CSF1 | 17230 | -0.000 | 0.0431 | No |
| 190 | SOCS1 | 17268 | -0.000 | 0.0410 | No |
| 191 | TLR7 | 17462 | -0.000 | 0.0302 | No |
| 192 | IGF2R | 17481 | -0.000 | 0.0292 | No |
| 193 | MYO1C | 17520 | -0.000 | 0.0271 | No |
| 194 | PRAF2 | 17802 | -0.000 | 0.0113 | No |