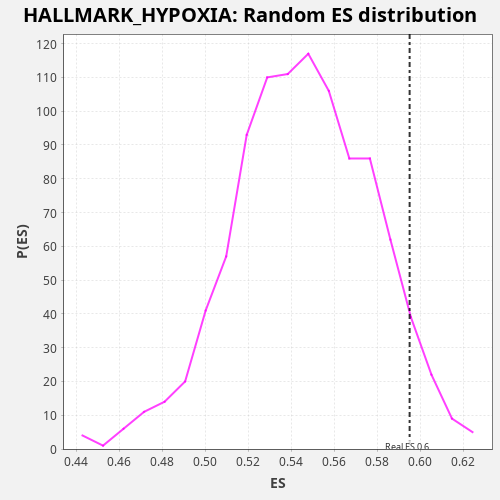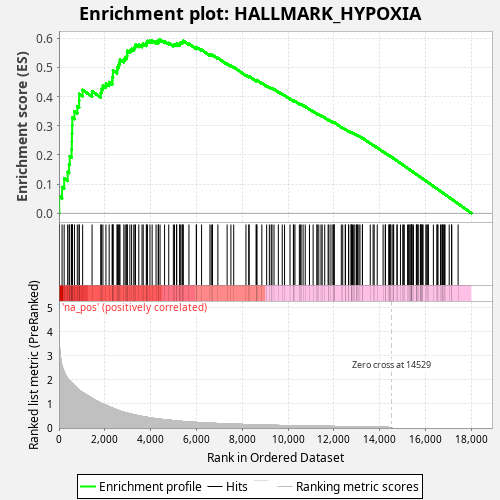
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_HYPOXIA |
| Enrichment Score (ES) | 0.5950715 |
| Normalized Enrichment Score (NES) | 1.0905259 |
| Nominal p-value | 0.05 |
| FDR q-value | 0.38735765 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | PDK1 | 8 | 3.901 | 0.0577 | Yes |
| 2 | PFKFB3 | 134 | 2.603 | 0.0895 | Yes |
| 3 | HS3ST1 | 225 | 2.326 | 0.1192 | Yes |
| 4 | MAFF | 378 | 2.060 | 0.1414 | Yes |
| 5 | CAVIN3 | 440 | 1.988 | 0.1676 | Yes |
| 6 | F3 | 467 | 1.956 | 0.1953 | Yes |
| 7 | PDK3 | 548 | 1.880 | 0.2189 | Yes |
| 8 | SDC3 | 560 | 1.871 | 0.2462 | Yes |
| 9 | SRPX | 565 | 1.865 | 0.2737 | Yes |
| 10 | CHST2 | 575 | 1.853 | 0.3009 | Yes |
| 11 | TPD52 | 581 | 1.850 | 0.3282 | Yes |
| 12 | NAGK | 674 | 1.759 | 0.3492 | Yes |
| 13 | BGN | 799 | 1.643 | 0.3668 | Yes |
| 14 | JMJD6 | 878 | 1.578 | 0.3860 | Yes |
| 15 | MT2A | 882 | 1.575 | 0.4093 | Yes |
| 16 | DDIT4 | 1032 | 1.475 | 0.4229 | Yes |
| 17 | SELENBP1 | 1443 | 1.233 | 0.4183 | Yes |
| 18 | B4GALNT2 | 1823 | 1.042 | 0.4125 | Yes |
| 19 | GRHPR | 1850 | 1.030 | 0.4264 | Yes |
| 20 | GALK1 | 1917 | 0.998 | 0.4376 | Yes |
| 21 | KLHL24 | 2049 | 0.946 | 0.4444 | Yes |
| 22 | CHST3 | 2189 | 0.885 | 0.4497 | Yes |
| 23 | UGP2 | 2324 | 0.831 | 0.4546 | Yes |
| 24 | GBE1 | 2328 | 0.829 | 0.4668 | Yes |
| 25 | PGM1 | 2357 | 0.818 | 0.4774 | Yes |
| 26 | ADORA2B | 2359 | 0.818 | 0.4896 | Yes |
| 27 | GYS1 | 2533 | 0.751 | 0.4911 | Yes |
| 28 | WSB1 | 2549 | 0.746 | 0.5014 | Yes |
| 29 | PKLR | 2602 | 0.728 | 0.5093 | Yes |
| 30 | DPYSL4 | 2633 | 0.713 | 0.5182 | Yes |
| 31 | FAM162A | 2663 | 0.704 | 0.5271 | Yes |
| 32 | PFKL | 2830 | 0.654 | 0.5275 | Yes |
| 33 | GAPDH | 2890 | 0.639 | 0.5338 | Yes |
| 34 | AMPD3 | 2959 | 0.621 | 0.5392 | Yes |
| 35 | TPBG | 2974 | 0.618 | 0.5476 | Yes |
| 36 | IGFBP1 | 2977 | 0.618 | 0.5567 | Yes |
| 37 | PYGM | 3094 | 0.589 | 0.5590 | Yes |
| 38 | ANKZF1 | 3170 | 0.572 | 0.5633 | Yes |
| 39 | NDST2 | 3265 | 0.548 | 0.5662 | Yes |
| 40 | LXN | 3316 | 0.540 | 0.5715 | Yes |
| 41 | LDHC | 3339 | 0.535 | 0.5782 | Yes |
| 42 | HEXA | 3484 | 0.506 | 0.5777 | Yes |
| 43 | KDELR3 | 3625 | 0.480 | 0.5770 | Yes |
| 44 | PGAM2 | 3676 | 0.469 | 0.5812 | Yes |
| 45 | ENO3 | 3811 | 0.447 | 0.5803 | Yes |
| 46 | STBD1 | 3829 | 0.445 | 0.5860 | Yes |
| 47 | PCK1 | 3861 | 0.440 | 0.5908 | Yes |
| 48 | GCNT2 | 3971 | 0.424 | 0.5910 | Yes |
| 49 | GAPDHS | 4066 | 0.411 | 0.5919 | Yes |
| 50 | FBP1 | 4237 | 0.386 | 0.5881 | Yes |
| 51 | CP | 4333 | 0.373 | 0.5883 | Yes |
| 52 | CA12 | 4338 | 0.373 | 0.5937 | Yes |
| 53 | VLDLR | 4411 | 0.366 | 0.5951 | Yes |
| 54 | ZNF292 | 4609 | 0.344 | 0.5891 | No |
| 55 | ALDOB | 4791 | 0.325 | 0.5838 | No |
| 56 | VEGFA | 4992 | 0.306 | 0.5772 | No |
| 57 | CASP6 | 5043 | 0.302 | 0.5789 | No |
| 58 | ERRFI1 | 5130 | 0.294 | 0.5784 | No |
| 59 | PPP1R15A | 5141 | 0.293 | 0.5822 | No |
| 60 | SCARB1 | 5270 | 0.281 | 0.5792 | No |
| 61 | PGM2 | 5276 | 0.281 | 0.5832 | No |
| 62 | BCAN | 5320 | 0.278 | 0.5849 | No |
| 63 | ILVBL | 5380 | 0.272 | 0.5856 | No |
| 64 | B3GALT6 | 5406 | 0.270 | 0.5883 | No |
| 65 | SLC25A1 | 5425 | 0.269 | 0.5913 | No |
| 66 | LDHA | 5673 | 0.251 | 0.5811 | No |
| 67 | SLC2A3 | 5992 | 0.232 | 0.5667 | No |
| 68 | PLIN2 | 6004 | 0.231 | 0.5696 | No |
| 69 | PHKG1 | 6224 | 0.220 | 0.5605 | No |
| 70 | ENO1 | 6590 | 0.201 | 0.5430 | No |
| 71 | SLC2A5 | 6648 | 0.198 | 0.5428 | No |
| 72 | MT1E | 6702 | 0.195 | 0.5427 | No |
| 73 | NCAN | 6936 | 0.186 | 0.5324 | No |
| 74 | CCN5 | 7340 | 0.169 | 0.5123 | No |
| 75 | NDRG1 | 7505 | 0.164 | 0.5055 | No |
| 76 | GPI | 7627 | 0.160 | 0.5011 | No |
| 77 | HMOX1 | 8162 | 0.142 | 0.4733 | No |
| 78 | PAM | 8283 | 0.138 | 0.4686 | No |
| 79 | TPI1 | 8301 | 0.137 | 0.4697 | No |
| 80 | NDST1 | 8609 | 0.129 | 0.4543 | No |
| 81 | SDC4 | 8614 | 0.129 | 0.4560 | No |
| 82 | NOCT | 8645 | 0.128 | 0.4563 | No |
| 83 | SULT2B1 | 8855 | 0.123 | 0.4464 | No |
| 84 | HK2 | 9065 | 0.118 | 0.4364 | No |
| 85 | CDKN1A | 9183 | 0.114 | 0.4315 | No |
| 86 | RORA | 9263 | 0.112 | 0.4288 | No |
| 87 | ALDOC | 9298 | 0.112 | 0.4285 | No |
| 88 | MIF | 9386 | 0.109 | 0.4253 | No |
| 89 | EGFR | 9576 | 0.106 | 0.4162 | No |
| 90 | ANXA2 | 9746 | 0.102 | 0.4082 | No |
| 91 | LARGE1 | 9847 | 0.100 | 0.4041 | No |
| 92 | ERO1A | 10085 | 0.095 | 0.3922 | No |
| 93 | TMEM45A | 10232 | 0.093 | 0.3854 | No |
| 94 | CCNG2 | 10249 | 0.092 | 0.3859 | No |
| 95 | AKAP12 | 10300 | 0.091 | 0.3845 | No |
| 96 | PPP1R3C | 10493 | 0.088 | 0.3750 | No |
| 97 | TGFBI | 10535 | 0.088 | 0.3740 | No |
| 98 | TPST2 | 10570 | 0.087 | 0.3734 | No |
| 99 | PFKP | 10656 | 0.085 | 0.3699 | No |
| 100 | MAP3K1 | 10750 | 0.084 | 0.3659 | No |
| 101 | PRKCA | 10934 | 0.080 | 0.3568 | No |
| 102 | SLC37A4 | 11094 | 0.077 | 0.3491 | No |
| 103 | MYH9 | 11259 | 0.075 | 0.3410 | No |
| 104 | TIPARP | 11271 | 0.075 | 0.3415 | No |
| 105 | ALDOA | 11342 | 0.074 | 0.3386 | No |
| 106 | CAVIN1 | 11438 | 0.073 | 0.3344 | No |
| 107 | PPARGC1A | 11489 | 0.072 | 0.3327 | No |
| 108 | CAV1 | 11600 | 0.070 | 0.3275 | No |
| 109 | XPNPEP1 | 11749 | 0.068 | 0.3202 | No |
| 110 | GLRX | 11783 | 0.067 | 0.3194 | No |
| 111 | HOXB9 | 11872 | 0.066 | 0.3154 | No |
| 112 | HK1 | 11964 | 0.064 | 0.3113 | No |
| 113 | IRS2 | 11971 | 0.064 | 0.3119 | No |
| 114 | PLAUR | 12009 | 0.064 | 0.3107 | No |
| 115 | ENO2 | 12012 | 0.064 | 0.3116 | No |
| 116 | GPC1 | 12324 | 0.060 | 0.2950 | No |
| 117 | ISG20 | 12387 | 0.059 | 0.2924 | No |
| 118 | GCK | 12497 | 0.057 | 0.2871 | No |
| 119 | NEDD4L | 12504 | 0.057 | 0.2876 | No |
| 120 | PNRC1 | 12643 | 0.055 | 0.2807 | No |
| 121 | AK4 | 12646 | 0.055 | 0.2814 | No |
| 122 | TGM2 | 12754 | 0.054 | 0.2762 | No |
| 123 | P4HA1 | 12758 | 0.053 | 0.2769 | No |
| 124 | DTNA | 12764 | 0.053 | 0.2774 | No |
| 125 | ATF3 | 12813 | 0.053 | 0.2755 | No |
| 126 | P4HA2 | 12865 | 0.052 | 0.2734 | No |
| 127 | IGFBP3 | 12916 | 0.051 | 0.2713 | No |
| 128 | DDIT3 | 12990 | 0.051 | 0.2680 | No |
| 129 | CSRP2 | 13007 | 0.050 | 0.2678 | No |
| 130 | PPFIA4 | 13028 | 0.050 | 0.2675 | No |
| 131 | HDLBP | 13090 | 0.049 | 0.2648 | No |
| 132 | MXI1 | 13139 | 0.048 | 0.2628 | No |
| 133 | SLC6A6 | 13244 | 0.047 | 0.2577 | No |
| 134 | INHA | 13249 | 0.047 | 0.2581 | No |
| 135 | KIF5A | 13588 | 0.042 | 0.2398 | No |
| 136 | HSPA5 | 13713 | 0.041 | 0.2334 | No |
| 137 | EFNA3 | 13759 | 0.040 | 0.2315 | No |
| 138 | NR3C1 | 13893 | 0.038 | 0.2246 | No |
| 139 | DUSP1 | 14149 | 0.033 | 0.2108 | No |
| 140 | PGK1 | 14242 | 0.031 | 0.2061 | No |
| 141 | TKTL1 | 14253 | 0.031 | 0.2060 | No |
| 142 | BTG1 | 14396 | 0.022 | 0.1983 | No |
| 143 | KLF6 | 14423 | 0.021 | 0.1972 | No |
| 144 | ATP7A | 14443 | 0.020 | 0.1964 | No |
| 145 | KLF7 | 14458 | 0.019 | 0.1959 | No |
| 146 | ADM | 14493 | 0.016 | 0.1942 | No |
| 147 | IDS | 14581 | -0.000 | 0.1894 | No |
| 148 | DCN | 14589 | -0.000 | 0.1890 | No |
| 149 | RRAGD | 14611 | -0.000 | 0.1878 | No |
| 150 | FOSL2 | 14755 | -0.000 | 0.1798 | No |
| 151 | GPC4 | 14764 | -0.000 | 0.1793 | No |
| 152 | PDGFB | 14907 | -0.000 | 0.1713 | No |
| 153 | BRS3 | 15006 | -0.000 | 0.1658 | No |
| 154 | BNIP3L | 15050 | -0.000 | 0.1634 | No |
| 155 | HAS1 | 15071 | -0.000 | 0.1623 | No |
| 156 | CDKN1B | 15217 | -0.000 | 0.1542 | No |
| 157 | LOX | 15264 | -0.000 | 0.1516 | No |
| 158 | STC2 | 15284 | -0.000 | 0.1505 | No |
| 159 | KDM3A | 15327 | -0.000 | 0.1481 | No |
| 160 | SLC2A1 | 15365 | -0.000 | 0.1461 | No |
| 161 | TNFAIP3 | 15390 | -0.000 | 0.1447 | No |
| 162 | CCN2 | 15391 | -0.000 | 0.1447 | No |
| 163 | FOXO3 | 15393 | -0.000 | 0.1447 | No |
| 164 | PGF | 15409 | -0.000 | 0.1438 | No |
| 165 | TGFB3 | 15412 | -0.000 | 0.1437 | No |
| 166 | CXCR4 | 15475 | -0.000 | 0.1402 | No |
| 167 | PRDX5 | 15600 | -0.000 | 0.1333 | No |
| 168 | EDN2 | 15612 | -0.000 | 0.1326 | No |
| 169 | ZFP36 | 15627 | -0.000 | 0.1319 | No |
| 170 | CDKN1C | 15664 | -0.000 | 0.1298 | No |
| 171 | COL5A1 | 15681 | -0.000 | 0.1289 | No |
| 172 | VHL | 15777 | -0.000 | 0.1236 | No |
| 173 | BHLHE40 | 15778 | -0.000 | 0.1236 | No |
| 174 | ETS1 | 15799 | -0.000 | 0.1225 | No |
| 175 | TES | 15810 | -0.000 | 0.1219 | No |
| 176 | IL6 | 15843 | -0.000 | 0.1201 | No |
| 177 | PIM1 | 15881 | -0.000 | 0.1180 | No |
| 178 | IER3 | 15886 | -0.000 | 0.1178 | No |
| 179 | CCN1 | 16014 | -0.000 | 0.1107 | No |
| 180 | ACKR3 | 16059 | -0.000 | 0.1082 | No |
| 181 | PLAC8 | 16078 | -0.000 | 0.1072 | No |
| 182 | GPC3 | 16128 | -0.000 | 0.1045 | No |
| 183 | STC1 | 16344 | -0.000 | 0.0924 | No |
| 184 | SAP30 | 16497 | -0.000 | 0.0838 | No |
| 185 | CITED2 | 16516 | -0.000 | 0.0828 | No |
| 186 | NFIL3 | 16546 | -0.000 | 0.0812 | No |
| 187 | LALBA | 16654 | -0.000 | 0.0752 | No |
| 188 | RBPJ | 16683 | -0.000 | 0.0736 | No |
| 189 | EFNA1 | 16735 | -0.000 | 0.0708 | No |
| 190 | SDC2 | 16745 | -0.000 | 0.0703 | No |
| 191 | FOS | 16780 | -0.000 | 0.0683 | No |
| 192 | GAA | 16828 | -0.000 | 0.0657 | No |
| 193 | BCL2 | 16852 | -0.000 | 0.0644 | No |
| 194 | JUN | 17035 | -0.000 | 0.0542 | No |
| 195 | SIAH2 | 17137 | -0.000 | 0.0485 | No |
| 196 | EXT1 | 17147 | -0.000 | 0.0480 | No |
| 197 | S100A4 | 17423 | -0.000 | 0.0326 | No |