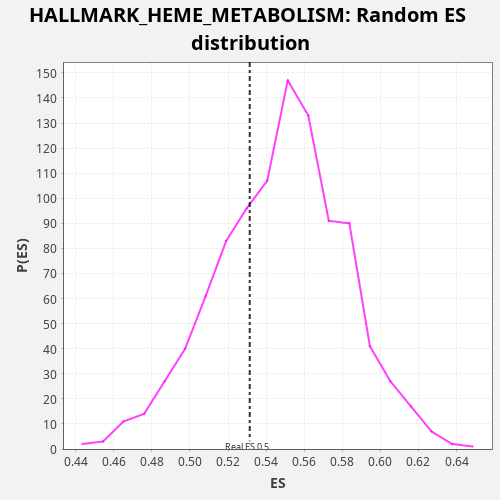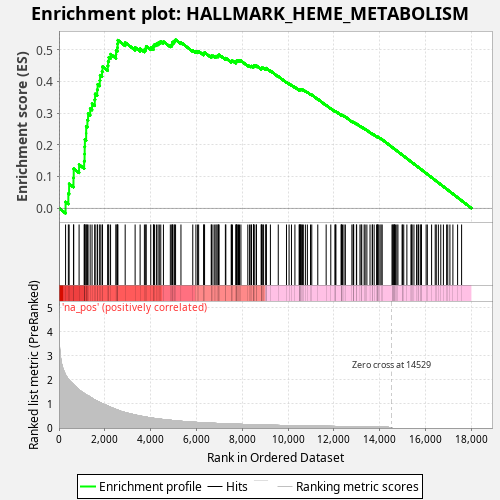
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_HEME_METABOLISM |
| Enrichment Score (ES) | 0.53137034 |
| Normalized Enrichment Score (NES) | 0.97001046 |
| Nominal p-value | 0.696 |
| FDR q-value | 0.8648193 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | SEC14L1 | 287 | 2.197 | 0.0199 | Yes |
| 2 | ALAD | 408 | 2.021 | 0.0463 | Yes |
| 3 | CCND3 | 442 | 1.984 | 0.0770 | Yes |
| 4 | CLCN3 | 632 | 1.796 | 0.0958 | Yes |
| 5 | HTRA2 | 643 | 1.786 | 0.1246 | Yes |
| 6 | DMTN | 877 | 1.579 | 0.1374 | Yes |
| 7 | HEBP1 | 1098 | 1.433 | 0.1485 | Yes |
| 8 | EZH1 | 1117 | 1.418 | 0.1708 | Yes |
| 9 | GAPVD1 | 1122 | 1.414 | 0.1937 | Yes |
| 10 | CTSB | 1131 | 1.409 | 0.2164 | Yes |
| 11 | RNF123 | 1185 | 1.377 | 0.2360 | Yes |
| 12 | OPTN | 1187 | 1.377 | 0.2585 | Yes |
| 13 | HTATIP2 | 1240 | 1.349 | 0.2777 | Yes |
| 14 | RCL1 | 1269 | 1.334 | 0.2980 | Yes |
| 15 | RIOK3 | 1360 | 1.280 | 0.3140 | Yes |
| 16 | SELENBP1 | 1443 | 1.233 | 0.3296 | Yes |
| 17 | BCAM | 1561 | 1.164 | 0.3421 | Yes |
| 18 | RHCE | 1575 | 1.159 | 0.3604 | Yes |
| 19 | SLC25A37 | 1667 | 1.112 | 0.3735 | Yes |
| 20 | BLVRA | 1687 | 1.104 | 0.3906 | Yes |
| 21 | NNT | 1780 | 1.062 | 0.4028 | Yes |
| 22 | UBAC1 | 1794 | 1.055 | 0.4194 | Yes |
| 23 | HBD | 1879 | 1.015 | 0.4313 | Yes |
| 24 | FN3K | 1900 | 1.005 | 0.4467 | Yes |
| 25 | LRP10 | 2127 | 0.914 | 0.4490 | Yes |
| 26 | TRIM10 | 2144 | 0.907 | 0.4629 | Yes |
| 27 | DAAM1 | 2174 | 0.891 | 0.4759 | Yes |
| 28 | SMOX | 2251 | 0.859 | 0.4857 | Yes |
| 29 | MOCOS | 2489 | 0.768 | 0.4850 | Yes |
| 30 | CDR2 | 2492 | 0.768 | 0.4975 | Yes |
| 31 | DCAF11 | 2551 | 0.745 | 0.5065 | Yes |
| 32 | SIDT2 | 2558 | 0.744 | 0.5183 | Yes |
| 33 | E2F2 | 2569 | 0.740 | 0.5299 | Yes |
| 34 | FECH | 2889 | 0.639 | 0.5224 | Yes |
| 35 | CAT | 3323 | 0.539 | 0.5070 | Yes |
| 36 | CTNS | 3541 | 0.496 | 0.5029 | Yes |
| 37 | RHAG | 3728 | 0.460 | 0.5000 | Yes |
| 38 | ACSL6 | 3785 | 0.452 | 0.5043 | Yes |
| 39 | RBM5 | 3806 | 0.449 | 0.5105 | Yes |
| 40 | SPTA1 | 4005 | 0.420 | 0.5063 | Yes |
| 41 | NARF | 4123 | 0.401 | 0.5063 | Yes |
| 42 | EPOR | 4127 | 0.401 | 0.5127 | Yes |
| 43 | OSBP2 | 4172 | 0.394 | 0.5167 | Yes |
| 44 | IGSF3 | 4253 | 0.384 | 0.5185 | Yes |
| 45 | GDE1 | 4322 | 0.374 | 0.5208 | Yes |
| 46 | HBZ | 4378 | 0.369 | 0.5238 | Yes |
| 47 | NCOA4 | 4443 | 0.361 | 0.5261 | Yes |
| 48 | CTSE | 4559 | 0.349 | 0.5254 | Yes |
| 49 | KLF1 | 4860 | 0.319 | 0.5138 | Yes |
| 50 | KHNYN | 4924 | 0.313 | 0.5154 | Yes |
| 51 | FTCD | 4943 | 0.311 | 0.5194 | Yes |
| 52 | PDZK1IP1 | 4954 | 0.310 | 0.5240 | Yes |
| 53 | TRAK2 | 5020 | 0.304 | 0.5253 | Yes |
| 54 | H4C3 | 5049 | 0.301 | 0.5287 | Yes |
| 55 | CAST | 5089 | 0.298 | 0.5314 | Yes |
| 56 | ABCG2 | 5324 | 0.278 | 0.5228 | No |
| 57 | RHD | 5840 | 0.240 | 0.4978 | No |
| 58 | CIR1 | 5977 | 0.232 | 0.4940 | No |
| 59 | GYPC | 6051 | 0.229 | 0.4936 | No |
| 60 | SLC66A2 | 6096 | 0.226 | 0.4949 | No |
| 61 | HAGH | 6314 | 0.215 | 0.4862 | No |
| 62 | BMP2K | 6328 | 0.214 | 0.4890 | No |
| 63 | PIGQ | 6355 | 0.213 | 0.4910 | No |
| 64 | SLC7A11 | 6649 | 0.198 | 0.4778 | No |
| 65 | SDCBP | 6653 | 0.198 | 0.4809 | No |
| 66 | ACP5 | 6721 | 0.194 | 0.4803 | No |
| 67 | ARL2BP | 6799 | 0.191 | 0.4791 | No |
| 68 | HBQ1 | 6853 | 0.189 | 0.4792 | No |
| 69 | BLVRB | 6901 | 0.187 | 0.4797 | No |
| 70 | ERMAP | 6952 | 0.185 | 0.4799 | No |
| 71 | ICAM4 | 6960 | 0.185 | 0.4825 | No |
| 72 | SLC22A4 | 6989 | 0.183 | 0.4840 | No |
| 73 | EIF2AK1 | 7263 | 0.172 | 0.4714 | No |
| 74 | RNF19A | 7283 | 0.171 | 0.4732 | No |
| 75 | ADD1 | 7518 | 0.163 | 0.4627 | No |
| 76 | GYPE | 7531 | 0.163 | 0.4647 | No |
| 77 | ALDH1L1 | 7572 | 0.161 | 0.4651 | No |
| 78 | BPGM | 7721 | 0.156 | 0.4594 | No |
| 79 | LMO2 | 7733 | 0.156 | 0.4613 | No |
| 80 | ENDOD1 | 7748 | 0.156 | 0.4631 | No |
| 81 | ACKR1 | 7757 | 0.155 | 0.4652 | No |
| 82 | PC | 7802 | 0.154 | 0.4652 | No |
| 83 | ARHGEF12 | 7836 | 0.153 | 0.4659 | No |
| 84 | SPTB | 7881 | 0.152 | 0.4659 | No |
| 85 | ASNS | 7942 | 0.149 | 0.4650 | No |
| 86 | GYPA | 8243 | 0.139 | 0.4504 | No |
| 87 | PGLS | 8320 | 0.137 | 0.4484 | No |
| 88 | ALDH6A1 | 8378 | 0.135 | 0.4474 | No |
| 89 | MFHAS1 | 8403 | 0.134 | 0.4483 | No |
| 90 | UCP2 | 8488 | 0.132 | 0.4457 | No |
| 91 | BTRC | 8493 | 0.132 | 0.4477 | No |
| 92 | SYNJ1 | 8502 | 0.132 | 0.4494 | No |
| 93 | KEL | 8520 | 0.131 | 0.4506 | No |
| 94 | LPIN2 | 8610 | 0.129 | 0.4477 | No |
| 95 | BACH1 | 8618 | 0.129 | 0.4494 | No |
| 96 | PSMD9 | 8825 | 0.124 | 0.4399 | No |
| 97 | SLC25A38 | 8839 | 0.123 | 0.4412 | No |
| 98 | KAT2B | 8850 | 0.123 | 0.4426 | No |
| 99 | MARK3 | 8894 | 0.122 | 0.4422 | No |
| 100 | ANK1 | 8932 | 0.121 | 0.4421 | No |
| 101 | BSG | 9030 | 0.119 | 0.4386 | No |
| 102 | ATG4A | 9041 | 0.118 | 0.4400 | No |
| 103 | PPOX | 9046 | 0.118 | 0.4417 | No |
| 104 | UROD | 9229 | 0.113 | 0.4333 | No |
| 105 | TMCC2 | 9572 | 0.106 | 0.4159 | No |
| 106 | TNRC6B | 9931 | 0.098 | 0.3974 | No |
| 107 | HBB | 10044 | 0.096 | 0.3927 | No |
| 108 | TFRC | 10149 | 0.094 | 0.3884 | No |
| 109 | TYR | 10299 | 0.092 | 0.3815 | No |
| 110 | SLC6A8 | 10483 | 0.089 | 0.3727 | No |
| 111 | FBXO9 | 10504 | 0.088 | 0.3730 | No |
| 112 | RBM38 | 10510 | 0.088 | 0.3742 | No |
| 113 | EPB41 | 10538 | 0.088 | 0.3741 | No |
| 114 | GLRX5 | 10559 | 0.087 | 0.3744 | No |
| 115 | BTG2 | 10578 | 0.087 | 0.3748 | No |
| 116 | TSPO2 | 10628 | 0.086 | 0.3735 | No |
| 117 | SLC30A10 | 10666 | 0.085 | 0.3728 | No |
| 118 | GYPB | 10756 | 0.084 | 0.3692 | No |
| 119 | FBXO7 | 10843 | 0.082 | 0.3657 | No |
| 120 | GCLC | 10980 | 0.080 | 0.3594 | No |
| 121 | MINPP1 | 11035 | 0.078 | 0.3576 | No |
| 122 | MGST3 | 11297 | 0.075 | 0.3442 | No |
| 123 | VEZF1 | 11663 | 0.069 | 0.3248 | No |
| 124 | UROS | 11865 | 0.066 | 0.3146 | No |
| 125 | CA1 | 12045 | 0.063 | 0.3056 | No |
| 126 | MYL4 | 12075 | 0.063 | 0.3050 | No |
| 127 | TNS1 | 12087 | 0.063 | 0.3054 | No |
| 128 | ADD2 | 12316 | 0.060 | 0.2936 | No |
| 129 | CPOX | 12330 | 0.060 | 0.2938 | No |
| 130 | CA2 | 12332 | 0.060 | 0.2948 | No |
| 131 | ATP6V0A1 | 12378 | 0.059 | 0.2932 | No |
| 132 | MBOAT2 | 12441 | 0.058 | 0.2907 | No |
| 133 | KDM7A | 12501 | 0.057 | 0.2883 | No |
| 134 | AGPAT4 | 12787 | 0.053 | 0.2732 | No |
| 135 | SLC6A9 | 12846 | 0.052 | 0.2708 | No |
| 136 | AQP3 | 12850 | 0.052 | 0.2714 | No |
| 137 | P4HA2 | 12865 | 0.052 | 0.2715 | No |
| 138 | TAL1 | 12983 | 0.051 | 0.2658 | No |
| 139 | PPP2R5B | 12994 | 0.051 | 0.2660 | No |
| 140 | MXI1 | 13139 | 0.048 | 0.2587 | No |
| 141 | RAD23A | 13191 | 0.048 | 0.2567 | No |
| 142 | NFE2 | 13217 | 0.047 | 0.2560 | No |
| 143 | SLC4A1 | 13321 | 0.046 | 0.2510 | No |
| 144 | NEK7 | 13364 | 0.046 | 0.2494 | No |
| 145 | TCEA1 | 13441 | 0.045 | 0.2459 | No |
| 146 | TSPAN5 | 13578 | 0.042 | 0.2389 | No |
| 147 | ADIPOR1 | 13675 | 0.041 | 0.2342 | No |
| 148 | GCLM | 13683 | 0.041 | 0.2345 | No |
| 149 | RANBP10 | 13758 | 0.040 | 0.2310 | No |
| 150 | SLC11A2 | 13865 | 0.039 | 0.2257 | No |
| 151 | NR3C1 | 13893 | 0.038 | 0.2248 | No |
| 152 | ELL2 | 13910 | 0.038 | 0.2245 | No |
| 153 | C3 | 13925 | 0.037 | 0.2243 | No |
| 154 | TFDP2 | 13932 | 0.037 | 0.2246 | No |
| 155 | HMBS | 14009 | 0.036 | 0.2209 | No |
| 156 | ABCB6 | 14061 | 0.035 | 0.2186 | No |
| 157 | EPB42 | 14109 | 0.034 | 0.2166 | No |
| 158 | CDC27 | 14542 | -0.000 | 0.1923 | No |
| 159 | LAMP2 | 14552 | -0.000 | 0.1918 | No |
| 160 | CCDC28A | 14610 | -0.000 | 0.1886 | No |
| 161 | MAP2K3 | 14622 | -0.000 | 0.1880 | No |
| 162 | DCUN1D1 | 14640 | -0.000 | 0.1870 | No |
| 163 | XK | 14647 | -0.000 | 0.1867 | No |
| 164 | FOXJ2 | 14687 | -0.000 | 0.1845 | No |
| 165 | PICALM | 14747 | -0.000 | 0.1812 | No |
| 166 | NFE2L1 | 14796 | -0.000 | 0.1785 | No |
| 167 | MOSPD1 | 14978 | -0.000 | 0.1683 | No |
| 168 | GATA1 | 15000 | -0.000 | 0.1672 | No |
| 169 | BNIP3L | 15050 | -0.000 | 0.1644 | No |
| 170 | KLF3 | 15187 | -0.000 | 0.1568 | No |
| 171 | SLC2A1 | 15365 | -0.000 | 0.1468 | No |
| 172 | FOXO3 | 15393 | -0.000 | 0.1453 | No |
| 173 | YPEL5 | 15416 | -0.000 | 0.1441 | No |
| 174 | DCAF10 | 15496 | -0.000 | 0.1396 | No |
| 175 | SLC10A3 | 15610 | -0.000 | 0.1333 | No |
| 176 | XPO7 | 15673 | -0.000 | 0.1298 | No |
| 177 | MPP1 | 15692 | -0.000 | 0.1288 | No |
| 178 | MKRN1 | 15769 | -0.000 | 0.1245 | No |
| 179 | ISCA1 | 15804 | -0.000 | 0.1226 | No |
| 180 | USP15 | 15824 | -0.000 | 0.1216 | No |
| 181 | HDGF | 16027 | -0.000 | 0.1102 | No |
| 182 | SNCA | 16080 | -0.000 | 0.1073 | No |
| 183 | CLIC2 | 16266 | -0.000 | 0.0969 | No |
| 184 | TRIM58 | 16424 | -0.000 | 0.0881 | No |
| 185 | GMPS | 16470 | -0.000 | 0.0856 | No |
| 186 | MARCHF8 | 16562 | -0.000 | 0.0805 | No |
| 187 | PRDX2 | 16667 | -0.000 | 0.0746 | No |
| 188 | SLC30A1 | 16782 | -0.000 | 0.0682 | No |
| 189 | NUDT4 | 16923 | -0.000 | 0.0604 | No |
| 190 | TMEM9B | 16971 | -0.000 | 0.0577 | No |
| 191 | FBXO34 | 17066 | -0.000 | 0.0524 | No |
| 192 | TENT5C | 17196 | -0.000 | 0.0452 | No |
| 193 | H1-0 | 17401 | -0.000 | 0.0337 | No |
| 194 | TOP1 | 17576 | -0.000 | 0.0240 | No |