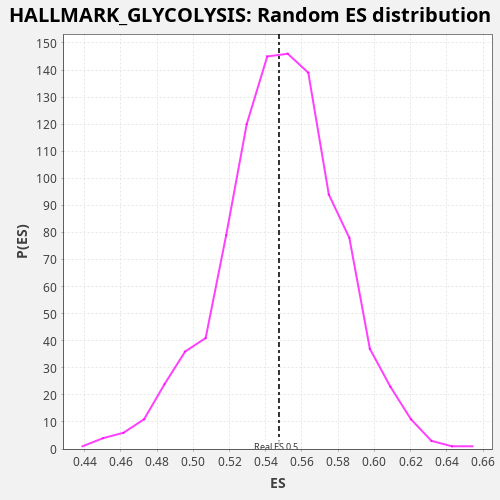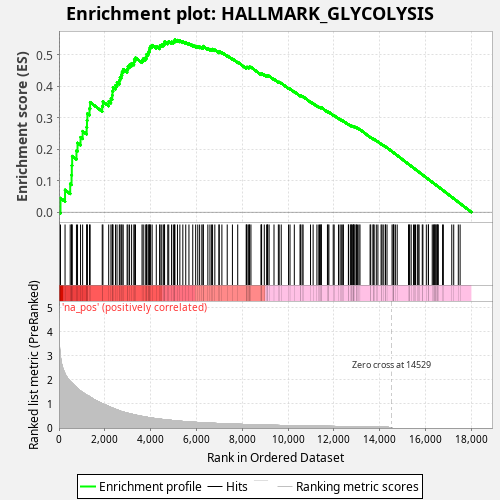
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_GLYCOLYSIS |
| Enrichment Score (ES) | 0.5473161 |
| Normalized Enrichment Score (NES) | 0.9979989 |
| Nominal p-value | 0.523 |
| FDR q-value | 0.87749946 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | GMPPA | 65 | 2.933 | 0.0448 | Yes |
| 2 | B3GAT1 | 264 | 2.240 | 0.0707 | Yes |
| 3 | GOT2 | 489 | 1.929 | 0.0899 | Yes |
| 4 | PDK3 | 548 | 1.880 | 0.1177 | Yes |
| 5 | SDC3 | 560 | 1.871 | 0.1480 | Yes |
| 6 | CHST2 | 575 | 1.853 | 0.1778 | Yes |
| 7 | GAL3ST1 | 762 | 1.687 | 0.1953 | Yes |
| 8 | MET | 809 | 1.633 | 0.2197 | Yes |
| 9 | B4GALT4 | 940 | 1.537 | 0.2377 | Yes |
| 10 | DDIT4 | 1032 | 1.475 | 0.2570 | Yes |
| 11 | GUSB | 1208 | 1.363 | 0.2697 | Yes |
| 12 | CLDN9 | 1222 | 1.357 | 0.2914 | Yes |
| 13 | GALE | 1238 | 1.351 | 0.3128 | Yes |
| 14 | SDC1 | 1332 | 1.302 | 0.3291 | Yes |
| 15 | CLN6 | 1359 | 1.280 | 0.3488 | Yes |
| 16 | RARS1 | 1887 | 1.012 | 0.3359 | Yes |
| 17 | GALK1 | 1917 | 0.998 | 0.3508 | Yes |
| 18 | DLD | 2169 | 0.894 | 0.3514 | Yes |
| 19 | B4GALT7 | 2268 | 0.853 | 0.3600 | Yes |
| 20 | UGP2 | 2324 | 0.831 | 0.3707 | Yes |
| 21 | FUT8 | 2334 | 0.827 | 0.3838 | Yes |
| 22 | ADORA2B | 2359 | 0.818 | 0.3960 | Yes |
| 23 | SPAG4 | 2468 | 0.776 | 0.4027 | Yes |
| 24 | GYS1 | 2533 | 0.751 | 0.4115 | Yes |
| 25 | DPYSL4 | 2633 | 0.713 | 0.4178 | Yes |
| 26 | FAM162A | 2663 | 0.704 | 0.4278 | Yes |
| 27 | CAPN5 | 2718 | 0.685 | 0.4360 | Yes |
| 28 | LCT | 2752 | 0.675 | 0.4453 | Yes |
| 29 | RBCK1 | 2803 | 0.660 | 0.4534 | Yes |
| 30 | TPBG | 2974 | 0.618 | 0.4541 | Yes |
| 31 | GNPDA1 | 3010 | 0.609 | 0.4622 | Yes |
| 32 | MED24 | 3081 | 0.592 | 0.4680 | Yes |
| 33 | ANKZF1 | 3170 | 0.572 | 0.4725 | Yes |
| 34 | TALDO1 | 3273 | 0.548 | 0.4758 | Yes |
| 35 | PYGB | 3284 | 0.546 | 0.4843 | Yes |
| 36 | LDHC | 3339 | 0.535 | 0.4901 | Yes |
| 37 | KDELR3 | 3625 | 0.480 | 0.4820 | Yes |
| 38 | PGAM2 | 3676 | 0.469 | 0.4870 | Yes |
| 39 | AGRN | 3766 | 0.454 | 0.4895 | Yes |
| 40 | NOL3 | 3818 | 0.446 | 0.4940 | Yes |
| 41 | ME2 | 3824 | 0.445 | 0.5011 | Yes |
| 42 | MDH2 | 3887 | 0.435 | 0.5048 | Yes |
| 43 | PGAM1 | 3920 | 0.431 | 0.5101 | Yes |
| 44 | AURKA | 3956 | 0.426 | 0.5151 | Yes |
| 45 | GPR87 | 3966 | 0.424 | 0.5216 | Yes |
| 46 | CACNA1H | 3999 | 0.421 | 0.5268 | Yes |
| 47 | GAPDHS | 4066 | 0.411 | 0.5299 | Yes |
| 48 | PMM2 | 4247 | 0.385 | 0.5261 | Yes |
| 49 | CHPF2 | 4393 | 0.368 | 0.5241 | Yes |
| 50 | VLDLR | 4411 | 0.366 | 0.5291 | Yes |
| 51 | BPNT1 | 4485 | 0.357 | 0.5309 | Yes |
| 52 | IDUA | 4555 | 0.349 | 0.5328 | Yes |
| 53 | PYGL | 4599 | 0.345 | 0.5361 | Yes |
| 54 | ZNF292 | 4609 | 0.344 | 0.5413 | Yes |
| 55 | CTH | 4749 | 0.329 | 0.5389 | Yes |
| 56 | ALDOB | 4791 | 0.325 | 0.5420 | Yes |
| 57 | QSOX1 | 4914 | 0.314 | 0.5403 | Yes |
| 58 | VEGFA | 4992 | 0.306 | 0.5411 | Yes |
| 59 | CASP6 | 5043 | 0.302 | 0.5433 | Yes |
| 60 | GYS2 | 5060 | 0.300 | 0.5473 | Yes |
| 61 | DEPDC1 | 5172 | 0.291 | 0.5459 | No |
| 62 | PGM2 | 5276 | 0.281 | 0.5447 | No |
| 63 | B3GALT6 | 5406 | 0.270 | 0.5420 | No |
| 64 | PLOD2 | 5533 | 0.261 | 0.5392 | No |
| 65 | LDHA | 5673 | 0.251 | 0.5355 | No |
| 66 | MIOX | 5839 | 0.240 | 0.5302 | No |
| 67 | AK3 | 5966 | 0.233 | 0.5270 | No |
| 68 | HS6ST2 | 6058 | 0.228 | 0.5257 | No |
| 69 | PKP2 | 6139 | 0.224 | 0.5249 | No |
| 70 | PLOD1 | 6235 | 0.219 | 0.5232 | No |
| 71 | SLC16A3 | 6293 | 0.216 | 0.5235 | No |
| 72 | ECD | 6298 | 0.216 | 0.5269 | No |
| 73 | EXT2 | 6498 | 0.205 | 0.5191 | No |
| 74 | ENO1 | 6590 | 0.201 | 0.5173 | No |
| 75 | LHPP | 6663 | 0.197 | 0.5165 | No |
| 76 | DSC2 | 6704 | 0.195 | 0.5175 | No |
| 77 | IDH1 | 6803 | 0.191 | 0.5151 | No |
| 78 | NDST3 | 6982 | 0.183 | 0.5082 | No |
| 79 | GNE | 6994 | 0.183 | 0.5106 | No |
| 80 | AKR1A1 | 7111 | 0.178 | 0.5070 | No |
| 81 | MPI | 7344 | 0.169 | 0.4968 | No |
| 82 | SLC25A10 | 7571 | 0.161 | 0.4867 | No |
| 83 | PC | 7802 | 0.154 | 0.4764 | No |
| 84 | MERTK | 8172 | 0.142 | 0.4580 | No |
| 85 | TPST1 | 8187 | 0.141 | 0.4595 | No |
| 86 | FBP2 | 8209 | 0.140 | 0.4607 | No |
| 87 | PAM | 8283 | 0.138 | 0.4589 | No |
| 88 | TPI1 | 8301 | 0.137 | 0.4602 | No |
| 89 | PGLS | 8320 | 0.137 | 0.4614 | No |
| 90 | PFKFB1 | 8379 | 0.135 | 0.4604 | No |
| 91 | GMPPB | 8817 | 0.124 | 0.4379 | No |
| 92 | COG2 | 8840 | 0.123 | 0.4387 | No |
| 93 | GOT1 | 8845 | 0.123 | 0.4405 | No |
| 94 | NT5E | 8962 | 0.120 | 0.4360 | No |
| 95 | HK2 | 9065 | 0.118 | 0.4322 | No |
| 96 | CHST12 | 9102 | 0.117 | 0.4321 | No |
| 97 | NDUFV3 | 9110 | 0.116 | 0.4336 | No |
| 98 | PKM | 9172 | 0.115 | 0.4321 | No |
| 99 | MIF | 9386 | 0.109 | 0.4220 | No |
| 100 | EGFR | 9576 | 0.106 | 0.4131 | No |
| 101 | XYLT2 | 9612 | 0.105 | 0.4129 | No |
| 102 | SRD5A3 | 9702 | 0.103 | 0.4096 | No |
| 103 | MDH1 | 10021 | 0.097 | 0.3933 | No |
| 104 | ERO1A | 10085 | 0.095 | 0.3913 | No |
| 105 | ALDH9A1 | 10273 | 0.092 | 0.3824 | No |
| 106 | ALG1 | 10528 | 0.088 | 0.3696 | No |
| 107 | TGFBI | 10535 | 0.088 | 0.3707 | No |
| 108 | NANP | 10611 | 0.086 | 0.3679 | No |
| 109 | PFKP | 10656 | 0.085 | 0.3668 | No |
| 110 | GCLC | 10980 | 0.080 | 0.3500 | No |
| 111 | SLC37A4 | 11094 | 0.077 | 0.3449 | No |
| 112 | FKBP4 | 11260 | 0.075 | 0.3369 | No |
| 113 | ALDOA | 11342 | 0.074 | 0.3336 | No |
| 114 | PSMC4 | 11373 | 0.074 | 0.3331 | No |
| 115 | NASP | 11425 | 0.073 | 0.3315 | No |
| 116 | GFPT1 | 11439 | 0.073 | 0.3319 | No |
| 117 | SDHC | 11452 | 0.072 | 0.3325 | No |
| 118 | GLCE | 11719 | 0.068 | 0.3186 | No |
| 119 | SOD1 | 11745 | 0.068 | 0.3184 | No |
| 120 | GLRX | 11783 | 0.067 | 0.3174 | No |
| 121 | IRS2 | 11971 | 0.064 | 0.3080 | No |
| 122 | ENO2 | 12012 | 0.064 | 0.3068 | No |
| 123 | VCAN | 12201 | 0.061 | 0.2972 | No |
| 124 | BIK | 12270 | 0.060 | 0.2944 | No |
| 125 | GPC1 | 12324 | 0.060 | 0.2924 | No |
| 126 | ISG20 | 12387 | 0.059 | 0.2899 | No |
| 127 | B3GNT3 | 12416 | 0.058 | 0.2893 | No |
| 128 | EGLN3 | 12634 | 0.055 | 0.2780 | No |
| 129 | AK4 | 12646 | 0.055 | 0.2783 | No |
| 130 | GALK2 | 12732 | 0.054 | 0.2744 | No |
| 131 | P4HA1 | 12758 | 0.053 | 0.2739 | No |
| 132 | LHX9 | 12759 | 0.053 | 0.2748 | No |
| 133 | ARTN | 12819 | 0.053 | 0.2723 | No |
| 134 | B4GALT2 | 12852 | 0.052 | 0.2714 | No |
| 135 | P4HA2 | 12865 | 0.052 | 0.2716 | No |
| 136 | TGFA | 12871 | 0.052 | 0.2722 | No |
| 137 | IGFBP3 | 12916 | 0.051 | 0.2706 | No |
| 138 | HMMR | 12976 | 0.051 | 0.2681 | No |
| 139 | TFF3 | 12984 | 0.051 | 0.2685 | No |
| 140 | PPFIA4 | 13028 | 0.050 | 0.2669 | No |
| 141 | HDLBP | 13090 | 0.049 | 0.2643 | No |
| 142 | MXI1 | 13139 | 0.048 | 0.2624 | No |
| 143 | PHKA2 | 13582 | 0.042 | 0.2383 | No |
| 144 | CYB5A | 13609 | 0.042 | 0.2375 | No |
| 145 | HSPA5 | 13713 | 0.041 | 0.2324 | No |
| 146 | B4GALT1 | 13714 | 0.041 | 0.2331 | No |
| 147 | EFNA3 | 13759 | 0.040 | 0.2313 | No |
| 148 | PPP2CB | 13852 | 0.039 | 0.2268 | No |
| 149 | SLC35A3 | 13922 | 0.038 | 0.2235 | No |
| 150 | ABCB6 | 14061 | 0.035 | 0.2164 | No |
| 151 | CHST1 | 14099 | 0.034 | 0.2148 | No |
| 152 | KIF2A | 14155 | 0.033 | 0.2123 | No |
| 153 | PGK1 | 14242 | 0.031 | 0.2080 | No |
| 154 | TKTL1 | 14253 | 0.031 | 0.2079 | No |
| 155 | HOMER1 | 14320 | 0.028 | 0.2047 | No |
| 156 | SLC25A13 | 14540 | -0.000 | 0.1924 | No |
| 157 | DCN | 14589 | -0.000 | 0.1897 | No |
| 158 | RRAGD | 14611 | -0.000 | 0.1885 | No |
| 159 | CD44 | 14614 | -0.000 | 0.1884 | No |
| 160 | ME1 | 14685 | -0.000 | 0.1845 | No |
| 161 | GPC4 | 14764 | -0.000 | 0.1801 | No |
| 162 | KIF20A | 15259 | -0.000 | 0.1524 | No |
| 163 | STC2 | 15284 | -0.000 | 0.1510 | No |
| 164 | CENPA | 15308 | -0.000 | 0.1497 | No |
| 165 | STMN1 | 15375 | -0.000 | 0.1460 | No |
| 166 | CXCR4 | 15475 | -0.000 | 0.1405 | No |
| 167 | CHPF | 15523 | -0.000 | 0.1378 | No |
| 168 | SOX9 | 15563 | -0.000 | 0.1356 | No |
| 169 | ARPP19 | 15643 | -0.000 | 0.1312 | No |
| 170 | COL5A1 | 15681 | -0.000 | 0.1291 | No |
| 171 | IL13RA1 | 15720 | -0.000 | 0.1270 | No |
| 172 | TXN | 15859 | -0.000 | 0.1192 | No |
| 173 | IER3 | 15886 | -0.000 | 0.1178 | No |
| 174 | HAX1 | 16036 | -0.000 | 0.1094 | No |
| 175 | PRPS1 | 16125 | -0.000 | 0.1045 | No |
| 176 | GPC3 | 16128 | -0.000 | 0.1044 | No |
| 177 | PAXIP1 | 16294 | -0.000 | 0.0951 | No |
| 178 | STC1 | 16344 | -0.000 | 0.0924 | No |
| 179 | G6PD | 16373 | -0.000 | 0.0908 | No |
| 180 | POLR3K | 16402 | -0.000 | 0.0892 | No |
| 181 | AGL | 16420 | -0.000 | 0.0883 | No |
| 182 | ELF3 | 16456 | -0.000 | 0.0863 | No |
| 183 | SAP30 | 16497 | -0.000 | 0.0840 | No |
| 184 | CITED2 | 16516 | -0.000 | 0.0830 | No |
| 185 | ALDH7A1 | 16537 | -0.000 | 0.0819 | No |
| 186 | CLDN3 | 16556 | -0.000 | 0.0809 | No |
| 187 | SDC2 | 16745 | -0.000 | 0.0703 | No |
| 188 | CDK1 | 16779 | -0.000 | 0.0685 | No |
| 189 | EXT1 | 17147 | -0.000 | 0.0479 | No |
| 190 | COPB2 | 17232 | -0.000 | 0.0432 | No |
| 191 | PPIA | 17432 | -0.000 | 0.0320 | No |
| 192 | RPE | 17511 | -0.000 | 0.0276 | No |