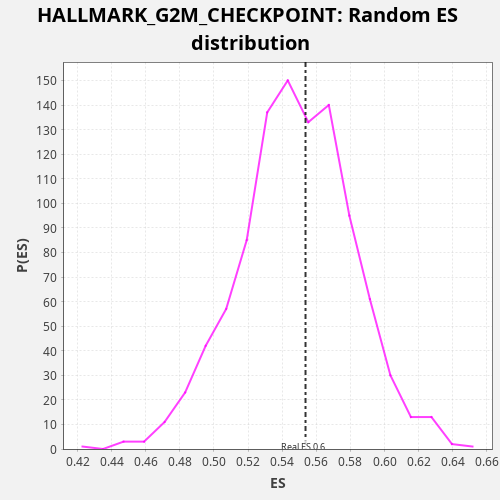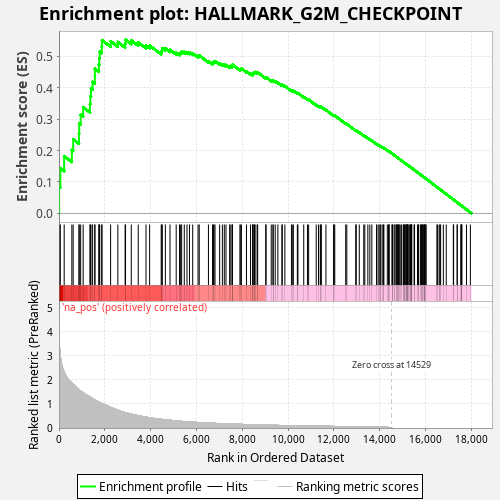
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_G2M_CHECKPOINT |
| Enrichment Score (ES) | 0.55367565 |
| Normalized Enrichment Score (NES) | 1.0098333 |
| Nominal p-value | 0.436 |
| FDR q-value | 0.8200716 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | MKI67 | 4 | 4.130 | 0.0847 | Yes |
| 2 | ATF5 | 58 | 3.017 | 0.1437 | Yes |
| 3 | TTK | 226 | 2.324 | 0.1822 | Yes |
| 4 | EXO1 | 561 | 1.870 | 0.2019 | Yes |
| 5 | PRMT5 | 618 | 1.814 | 0.2360 | Yes |
| 6 | DTYMK | 871 | 1.585 | 0.2544 | Yes |
| 7 | MT2A | 882 | 1.575 | 0.2863 | Yes |
| 8 | WRN | 946 | 1.534 | 0.3143 | Yes |
| 9 | H2BC12 | 1054 | 1.462 | 0.3383 | Yes |
| 10 | CENPE | 1352 | 1.284 | 0.3481 | Yes |
| 11 | CDC6 | 1368 | 1.274 | 0.3734 | Yes |
| 12 | SLC12A2 | 1397 | 1.260 | 0.3977 | Yes |
| 13 | TROAP | 1467 | 1.219 | 0.4189 | Yes |
| 14 | RACGAP1 | 1565 | 1.163 | 0.4374 | Yes |
| 15 | PDS5B | 1567 | 1.162 | 0.4612 | Yes |
| 16 | POLQ | 1738 | 1.079 | 0.4739 | Yes |
| 17 | KNL1 | 1756 | 1.072 | 0.4950 | Yes |
| 18 | CCNT1 | 1777 | 1.064 | 0.5157 | Yes |
| 19 | HUS1 | 1866 | 1.020 | 0.5317 | Yes |
| 20 | DBF4 | 1876 | 1.017 | 0.5521 | Yes |
| 21 | KIF5B | 2253 | 0.858 | 0.5487 | Yes |
| 22 | E2F2 | 2569 | 0.740 | 0.5462 | Yes |
| 23 | SMC4 | 2886 | 0.640 | 0.5416 | Yes |
| 24 | CHEK1 | 2905 | 0.636 | 0.5537 | Yes |
| 25 | MCM3 | 3155 | 0.574 | 0.5515 | No |
| 26 | EGF | 3459 | 0.512 | 0.5450 | No |
| 27 | ORC6 | 3798 | 0.450 | 0.5353 | No |
| 28 | AURKA | 3956 | 0.426 | 0.5352 | No |
| 29 | BRCA2 | 4461 | 0.360 | 0.5143 | No |
| 30 | STIL | 4494 | 0.356 | 0.5199 | No |
| 31 | TACC3 | 4506 | 0.354 | 0.5265 | No |
| 32 | RAD54L | 4643 | 0.341 | 0.5259 | No |
| 33 | TMPO | 4846 | 0.320 | 0.5211 | No |
| 34 | UBE2C | 5116 | 0.296 | 0.5121 | No |
| 35 | AURKB | 5261 | 0.282 | 0.5099 | No |
| 36 | HNRNPU | 5311 | 0.279 | 0.5128 | No |
| 37 | PRIM2 | 5354 | 0.274 | 0.5161 | No |
| 38 | ARID4A | 5469 | 0.266 | 0.5152 | No |
| 39 | TOP2A | 5588 | 0.258 | 0.5139 | No |
| 40 | ORC5 | 5696 | 0.249 | 0.5130 | No |
| 41 | KIF22 | 5837 | 0.240 | 0.5101 | No |
| 42 | TRAIP | 6070 | 0.228 | 0.5017 | No |
| 43 | PBK | 6122 | 0.225 | 0.5035 | No |
| 44 | PLK4 | 6524 | 0.204 | 0.4852 | No |
| 45 | TENT4A | 6703 | 0.195 | 0.4792 | No |
| 46 | CCNB2 | 6741 | 0.193 | 0.4811 | No |
| 47 | NSD2 | 6766 | 0.193 | 0.4837 | No |
| 48 | CDC45 | 6811 | 0.190 | 0.4851 | No |
| 49 | BUB1 | 7013 | 0.182 | 0.4776 | No |
| 50 | NEK2 | 7132 | 0.177 | 0.4746 | No |
| 51 | KIF15 | 7216 | 0.174 | 0.4735 | No |
| 52 | POLA2 | 7290 | 0.171 | 0.4729 | No |
| 53 | BIRC5 | 7452 | 0.165 | 0.4673 | No |
| 54 | MCM2 | 7471 | 0.165 | 0.4697 | No |
| 55 | CDC7 | 7558 | 0.162 | 0.4682 | No |
| 56 | KATNA1 | 7559 | 0.162 | 0.4715 | No |
| 57 | FANCC | 7573 | 0.161 | 0.4741 | No |
| 58 | GINS2 | 7898 | 0.151 | 0.4590 | No |
| 59 | KIF2C | 7937 | 0.149 | 0.4600 | No |
| 60 | MCM5 | 7963 | 0.149 | 0.4616 | No |
| 61 | JPT1 | 8184 | 0.141 | 0.4522 | No |
| 62 | POLE | 8361 | 0.136 | 0.4451 | No |
| 63 | ODF2 | 8455 | 0.133 | 0.4426 | No |
| 64 | DDX39A | 8457 | 0.133 | 0.4453 | No |
| 65 | NOLC1 | 8470 | 0.133 | 0.4473 | No |
| 66 | SMC2 | 8517 | 0.132 | 0.4474 | No |
| 67 | ESPL1 | 8535 | 0.131 | 0.4492 | No |
| 68 | BARD1 | 8562 | 0.130 | 0.4504 | No |
| 69 | LIG3 | 8648 | 0.128 | 0.4483 | No |
| 70 | CDKN3 | 8673 | 0.127 | 0.4495 | No |
| 71 | HOXC10 | 9026 | 0.119 | 0.4322 | No |
| 72 | CDC25B | 9039 | 0.118 | 0.4340 | No |
| 73 | SLC7A5 | 9265 | 0.112 | 0.4236 | No |
| 74 | EWSR1 | 9345 | 0.110 | 0.4215 | No |
| 75 | RASAL2 | 9347 | 0.110 | 0.4237 | No |
| 76 | KIF20B | 9439 | 0.108 | 0.4208 | No |
| 77 | NDC80 | 9555 | 0.106 | 0.4165 | No |
| 78 | NUP50 | 9732 | 0.102 | 0.4088 | No |
| 79 | MAP3K20 | 9745 | 0.102 | 0.4102 | No |
| 80 | HIF1A | 9861 | 0.099 | 0.4058 | No |
| 81 | NUSAP1 | 10147 | 0.094 | 0.3917 | No |
| 82 | KPNA2 | 10205 | 0.093 | 0.3904 | No |
| 83 | MCM6 | 10229 | 0.093 | 0.3910 | No |
| 84 | ODC1 | 10405 | 0.090 | 0.3831 | No |
| 85 | PTTG1 | 10431 | 0.090 | 0.3835 | No |
| 86 | MNAT1 | 10684 | 0.085 | 0.3711 | No |
| 87 | KPNB1 | 10847 | 0.082 | 0.3637 | No |
| 88 | RPA2 | 10898 | 0.081 | 0.3626 | No |
| 89 | MYBL2 | 11229 | 0.076 | 0.3456 | No |
| 90 | PLK1 | 11329 | 0.074 | 0.3416 | No |
| 91 | NCL | 11405 | 0.073 | 0.3389 | No |
| 92 | NASP | 11425 | 0.073 | 0.3393 | No |
| 93 | CCNA2 | 11454 | 0.072 | 0.3392 | No |
| 94 | LBR | 11653 | 0.069 | 0.3295 | No |
| 95 | LMNB1 | 11987 | 0.064 | 0.3121 | No |
| 96 | NOTCH2 | 12003 | 0.064 | 0.3126 | No |
| 97 | CASP8AP2 | 12039 | 0.063 | 0.3119 | No |
| 98 | G3BP1 | 12517 | 0.057 | 0.2863 | No |
| 99 | MAD2L1 | 12564 | 0.056 | 0.2849 | No |
| 100 | SMARCC1 | 12954 | 0.051 | 0.2641 | No |
| 101 | HMMR | 12976 | 0.051 | 0.2640 | No |
| 102 | CENPF | 13109 | 0.049 | 0.2576 | No |
| 103 | PRPF4B | 13304 | 0.047 | 0.2476 | No |
| 104 | FOXN3 | 13350 | 0.046 | 0.2460 | No |
| 105 | GSPT1 | 13483 | 0.044 | 0.2395 | No |
| 106 | AMD1 | 13570 | 0.042 | 0.2356 | No |
| 107 | E2F4 | 13657 | 0.041 | 0.2316 | No |
| 108 | RPS6KA5 | 13862 | 0.039 | 0.2210 | No |
| 109 | TPX2 | 13929 | 0.037 | 0.2180 | No |
| 110 | CDC25A | 13978 | 0.036 | 0.2161 | No |
| 111 | SS18 | 14025 | 0.036 | 0.2142 | No |
| 112 | BUB3 | 14054 | 0.035 | 0.2134 | No |
| 113 | KIF23 | 14136 | 0.033 | 0.2095 | No |
| 114 | SNRPD1 | 14172 | 0.033 | 0.2082 | No |
| 115 | TNPO2 | 14180 | 0.033 | 0.2085 | No |
| 116 | H2AZ1 | 14333 | 0.027 | 0.2005 | No |
| 117 | MEIS1 | 14384 | 0.023 | 0.1982 | No |
| 118 | SUV39H1 | 14402 | 0.022 | 0.1977 | No |
| 119 | HNRNPD | 14414 | 0.021 | 0.1975 | No |
| 120 | CDC27 | 14542 | -0.000 | 0.1904 | No |
| 121 | UPF1 | 14544 | -0.000 | 0.1903 | No |
| 122 | PAFAH1B1 | 14561 | -0.000 | 0.1894 | No |
| 123 | CUL3 | 14628 | -0.000 | 0.1857 | No |
| 124 | CUL1 | 14665 | -0.000 | 0.1837 | No |
| 125 | BCL3 | 14715 | -0.000 | 0.1810 | No |
| 126 | SMC1A | 14739 | -0.000 | 0.1797 | No |
| 127 | RBL1 | 14787 | -0.000 | 0.1770 | No |
| 128 | XPO1 | 14798 | -0.000 | 0.1765 | No |
| 129 | YTHDC1 | 14804 | -0.000 | 0.1762 | No |
| 130 | ATRX | 14811 | -0.000 | 0.1759 | No |
| 131 | KIF4A | 14856 | -0.000 | 0.1734 | No |
| 132 | HIRA | 14895 | -0.000 | 0.1713 | No |
| 133 | E2F1 | 14956 | -0.000 | 0.1679 | No |
| 134 | CTCF | 15028 | -0.000 | 0.1639 | No |
| 135 | SQLE | 15045 | -0.000 | 0.1630 | No |
| 136 | TGFB1 | 15066 | -0.000 | 0.1619 | No |
| 137 | H2AZ2 | 15091 | -0.000 | 0.1605 | No |
| 138 | EZH2 | 15104 | -0.000 | 0.1599 | No |
| 139 | UBE2S | 15144 | -0.000 | 0.1577 | No |
| 140 | CBX1 | 15153 | -0.000 | 0.1572 | No |
| 141 | HSPA8 | 15189 | -0.000 | 0.1553 | No |
| 142 | CCND1 | 15191 | -0.000 | 0.1552 | No |
| 143 | NUP98 | 15202 | -0.000 | 0.1546 | No |
| 144 | CDKN1B | 15217 | -0.000 | 0.1539 | No |
| 145 | SLC38A1 | 15219 | -0.000 | 0.1538 | No |
| 146 | MAPK14 | 15236 | -0.000 | 0.1529 | No |
| 147 | E2F3 | 15239 | -0.000 | 0.1528 | No |
| 148 | CENPA | 15308 | -0.000 | 0.1490 | No |
| 149 | SFPQ | 15345 | -0.000 | 0.1470 | No |
| 150 | CDC20 | 15366 | -0.000 | 0.1458 | No |
| 151 | DR1 | 15367 | -0.000 | 0.1458 | No |
| 152 | STMN1 | 15375 | -0.000 | 0.1454 | No |
| 153 | STAG1 | 15378 | -0.000 | 0.1453 | No |
| 154 | RAD23B | 15402 | -0.000 | 0.1440 | No |
| 155 | CDKN2C | 15501 | -0.000 | 0.1385 | No |
| 156 | CKS2 | 15522 | -0.000 | 0.1374 | No |
| 157 | ILF3 | 15652 | -0.000 | 0.1302 | No |
| 158 | DKC1 | 15691 | -0.000 | 0.1280 | No |
| 159 | CHMP1A | 15701 | -0.000 | 0.1275 | No |
| 160 | MEIS2 | 15779 | -0.000 | 0.1232 | No |
| 161 | SYNCRIP | 15813 | -0.000 | 0.1214 | No |
| 162 | CDK4 | 15820 | -0.000 | 0.1210 | No |
| 163 | TRA2B | 15854 | -0.000 | 0.1192 | No |
| 164 | MYC | 15873 | -0.000 | 0.1182 | No |
| 165 | KIF11 | 15907 | -0.000 | 0.1163 | No |
| 166 | SLC7A1 | 15940 | -0.000 | 0.1145 | No |
| 167 | CUL4A | 15953 | -0.000 | 0.1138 | No |
| 168 | TLE3 | 15969 | -0.000 | 0.1130 | No |
| 169 | PML | 15974 | -0.000 | 0.1128 | No |
| 170 | MTF2 | 16017 | -0.000 | 0.1104 | No |
| 171 | UCK2 | 16021 | -0.000 | 0.1102 | No |
| 172 | SAP30 | 16497 | -0.000 | 0.0836 | No |
| 173 | RAD21 | 16529 | -0.000 | 0.0818 | No |
| 174 | CUL5 | 16597 | -0.000 | 0.0781 | No |
| 175 | SMAD3 | 16632 | -0.000 | 0.0762 | No |
| 176 | CHAF1A | 16660 | -0.000 | 0.0747 | No |
| 177 | CDK1 | 16779 | -0.000 | 0.0680 | No |
| 178 | CKS1B | 16905 | -0.000 | 0.0610 | No |
| 179 | KMT5A | 17215 | -0.000 | 0.0437 | No |
| 180 | EFNA5 | 17227 | -0.000 | 0.0431 | No |
| 181 | H2AX | 17383 | -0.000 | 0.0344 | No |
| 182 | SRSF10 | 17385 | -0.000 | 0.0343 | No |
| 183 | TFDP1 | 17538 | -0.000 | 0.0258 | No |
| 184 | HMGN2 | 17573 | -0.000 | 0.0239 | No |
| 185 | TOP1 | 17576 | -0.000 | 0.0237 | No |
| 186 | PRC1 | 17577 | -0.000 | 0.0237 | No |
| 187 | DMD | 17583 | -0.000 | 0.0235 | No |
| 188 | RBM14 | 17789 | -0.000 | 0.0120 | No |
| 189 | MARCKS | 17959 | -0.000 | 0.0025 | No |