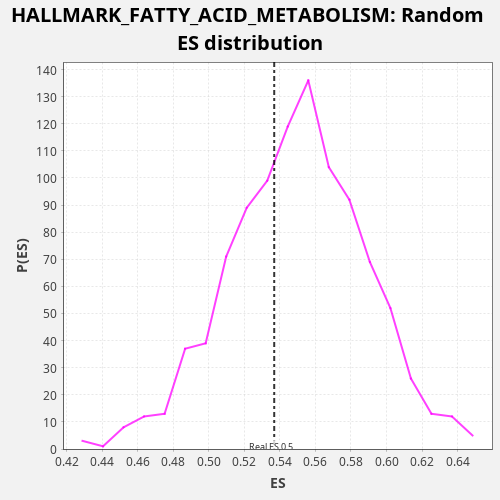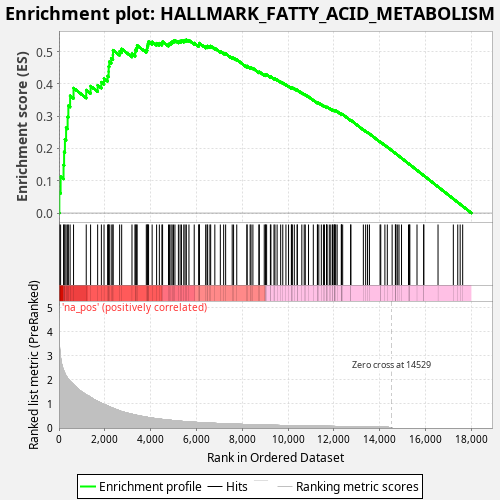
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | 0.53680843 |
| Normalized Enrichment Score (NES) | 0.9769741 |
| Nominal p-value | 0.645 |
| FDR q-value | 0.8419205 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | MGLL | 15 | 3.569 | 0.0635 | Yes |
| 2 | AUH | 70 | 2.902 | 0.1127 | Yes |
| 3 | ETFDH | 199 | 2.396 | 0.1487 | Yes |
| 4 | PPARA | 223 | 2.333 | 0.1895 | Yes |
| 5 | PDHA1 | 259 | 2.251 | 0.2281 | Yes |
| 6 | PTPRG | 310 | 2.164 | 0.2643 | Yes |
| 7 | TDO2 | 377 | 2.061 | 0.2977 | Yes |
| 8 | ALAD | 408 | 2.021 | 0.3325 | Yes |
| 9 | IDI1 | 485 | 1.934 | 0.3631 | Yes |
| 10 | ELOVL5 | 635 | 1.791 | 0.3870 | Yes |
| 11 | MLYCD | 1189 | 1.376 | 0.3808 | Yes |
| 12 | HPGD | 1379 | 1.269 | 0.3931 | Yes |
| 13 | BLVRA | 1687 | 1.104 | 0.3958 | Yes |
| 14 | GRHPR | 1850 | 1.030 | 0.4053 | Yes |
| 15 | PSME1 | 1966 | 0.977 | 0.4164 | Yes |
| 16 | NBN | 2122 | 0.915 | 0.4242 | Yes |
| 17 | FH | 2167 | 0.894 | 0.4379 | Yes |
| 18 | DLD | 2169 | 0.894 | 0.4539 | Yes |
| 19 | TP53INP2 | 2194 | 0.884 | 0.4685 | Yes |
| 20 | CIDEA | 2279 | 0.847 | 0.4791 | Yes |
| 21 | CRAT | 2354 | 0.819 | 0.4897 | Yes |
| 22 | RAP1GDS1 | 2358 | 0.818 | 0.5042 | Yes |
| 23 | HIBCH | 2647 | 0.711 | 0.5009 | Yes |
| 24 | ADH7 | 2736 | 0.679 | 0.5082 | Yes |
| 25 | SUCLG1 | 3186 | 0.570 | 0.4934 | Yes |
| 26 | BCKDHB | 3325 | 0.538 | 0.4953 | Yes |
| 27 | FMO1 | 3326 | 0.538 | 0.5050 | Yes |
| 28 | NCAPH2 | 3388 | 0.526 | 0.5111 | Yes |
| 29 | GSTZ1 | 3410 | 0.521 | 0.5193 | Yes |
| 30 | ENO3 | 3811 | 0.447 | 0.5049 | Yes |
| 31 | PRDX6 | 3849 | 0.442 | 0.5108 | Yes |
| 32 | GCDH | 3855 | 0.442 | 0.5185 | Yes |
| 33 | MDH2 | 3887 | 0.435 | 0.5246 | Yes |
| 34 | ACOT8 | 3910 | 0.432 | 0.5312 | Yes |
| 35 | GAPDHS | 4066 | 0.411 | 0.5299 | Yes |
| 36 | HSD17B4 | 4261 | 0.382 | 0.5259 | Yes |
| 37 | ACSL5 | 4381 | 0.369 | 0.5259 | Yes |
| 38 | CYP4A22 | 4493 | 0.356 | 0.5261 | Yes |
| 39 | ALDH3A2 | 4522 | 0.353 | 0.5309 | Yes |
| 40 | SUCLG2 | 4781 | 0.327 | 0.5223 | Yes |
| 41 | LTC4S | 4827 | 0.322 | 0.5256 | Yes |
| 42 | APEX1 | 4888 | 0.316 | 0.5280 | Yes |
| 43 | MCEE | 4940 | 0.311 | 0.5307 | Yes |
| 44 | AOC3 | 4997 | 0.306 | 0.5331 | Yes |
| 45 | GPD1 | 5061 | 0.300 | 0.5349 | Yes |
| 46 | ACADS | 5212 | 0.287 | 0.5317 | Yes |
| 47 | HMGCL | 5286 | 0.281 | 0.5327 | Yes |
| 48 | ACADM | 5346 | 0.275 | 0.5343 | Yes |
| 49 | ACAA2 | 5448 | 0.267 | 0.5335 | Yes |
| 50 | HAO2 | 5503 | 0.263 | 0.5352 | Yes |
| 51 | ACAT2 | 5559 | 0.260 | 0.5368 | Yes |
| 52 | LDHA | 5673 | 0.251 | 0.5350 | No |
| 53 | DECR1 | 5903 | 0.236 | 0.5264 | No |
| 54 | HADHB | 6099 | 0.226 | 0.5196 | No |
| 55 | CEL | 6121 | 0.225 | 0.5225 | No |
| 56 | IL4I1 | 6127 | 0.225 | 0.5262 | No |
| 57 | CRYZ | 6403 | 0.210 | 0.5146 | No |
| 58 | ALDH3A1 | 6462 | 0.207 | 0.5151 | No |
| 59 | VNN1 | 6491 | 0.206 | 0.5172 | No |
| 60 | BPHL | 6589 | 0.201 | 0.5154 | No |
| 61 | CD36 | 6622 | 0.199 | 0.5172 | No |
| 62 | IDH1 | 6803 | 0.191 | 0.5106 | No |
| 63 | RETSAT | 7043 | 0.181 | 0.5004 | No |
| 64 | ECHS1 | 7188 | 0.175 | 0.4955 | No |
| 65 | ACSS1 | 7275 | 0.172 | 0.4938 | No |
| 66 | CPT1A | 7561 | 0.162 | 0.4808 | No |
| 67 | CYP4A11 | 7621 | 0.160 | 0.4803 | No |
| 68 | PCBD1 | 7760 | 0.155 | 0.4754 | No |
| 69 | ACOT2 | 8204 | 0.141 | 0.4531 | No |
| 70 | CPT2 | 8210 | 0.140 | 0.4554 | No |
| 71 | ACAA1 | 8344 | 0.136 | 0.4504 | No |
| 72 | ACADL | 8383 | 0.135 | 0.4507 | No |
| 73 | UGDH | 8465 | 0.133 | 0.4485 | No |
| 74 | ACOX1 | 8727 | 0.126 | 0.4362 | No |
| 75 | ADSL | 8743 | 0.126 | 0.4376 | No |
| 76 | SUCLA2 | 8960 | 0.120 | 0.4277 | No |
| 77 | HSD17B7 | 9011 | 0.119 | 0.4270 | No |
| 78 | D2HGDH | 9025 | 0.119 | 0.4284 | No |
| 79 | UBE2L6 | 9064 | 0.118 | 0.4284 | No |
| 80 | UROD | 9229 | 0.113 | 0.4213 | No |
| 81 | HSDL2 | 9251 | 0.112 | 0.4221 | No |
| 82 | MIF | 9386 | 0.109 | 0.4166 | No |
| 83 | RDH16 | 9427 | 0.109 | 0.4163 | No |
| 84 | ADH1C | 9522 | 0.107 | 0.4130 | No |
| 85 | ACADVL | 9677 | 0.104 | 0.4062 | No |
| 86 | ECI2 | 9769 | 0.101 | 0.4029 | No |
| 87 | OSTC | 9907 | 0.099 | 0.3970 | No |
| 88 | MDH1 | 10021 | 0.097 | 0.3925 | No |
| 89 | RDH11 | 10144 | 0.094 | 0.3873 | No |
| 90 | AADAT | 10157 | 0.094 | 0.3883 | No |
| 91 | LGALS1 | 10187 | 0.094 | 0.3884 | No |
| 92 | ALDH9A1 | 10273 | 0.092 | 0.3853 | No |
| 93 | AQP7 | 10397 | 0.090 | 0.3800 | No |
| 94 | ODC1 | 10405 | 0.090 | 0.3813 | No |
| 95 | GABARAPL1 | 10601 | 0.086 | 0.3719 | No |
| 96 | HMGCS2 | 10704 | 0.085 | 0.3677 | No |
| 97 | SERINC1 | 10751 | 0.084 | 0.3666 | No |
| 98 | HSPH1 | 10893 | 0.081 | 0.3602 | No |
| 99 | GLUL | 11102 | 0.077 | 0.3500 | No |
| 100 | CD1D | 11281 | 0.075 | 0.3413 | No |
| 101 | PTS | 11284 | 0.075 | 0.3426 | No |
| 102 | ALDOA | 11342 | 0.074 | 0.3407 | No |
| 103 | SDHC | 11452 | 0.072 | 0.3359 | No |
| 104 | DHCR24 | 11545 | 0.071 | 0.3320 | No |
| 105 | PDHB | 11589 | 0.070 | 0.3309 | No |
| 106 | ACSL1 | 11679 | 0.069 | 0.3271 | No |
| 107 | DLST | 11680 | 0.069 | 0.3284 | No |
| 108 | CYP1A1 | 11706 | 0.068 | 0.3282 | No |
| 109 | CA4 | 11800 | 0.067 | 0.3242 | No |
| 110 | UROS | 11865 | 0.066 | 0.3218 | No |
| 111 | CA6 | 11939 | 0.065 | 0.3189 | No |
| 112 | ACO2 | 11958 | 0.064 | 0.3190 | No |
| 113 | ENO2 | 12012 | 0.064 | 0.3172 | No |
| 114 | CBR3 | 12031 | 0.063 | 0.3173 | No |
| 115 | EHHADH | 12046 | 0.063 | 0.3177 | No |
| 116 | SDHA | 12080 | 0.063 | 0.3170 | No |
| 117 | METAP1 | 12143 | 0.062 | 0.3146 | No |
| 118 | HSD17B11 | 12326 | 0.060 | 0.3055 | No |
| 119 | CPOX | 12330 | 0.060 | 0.3064 | No |
| 120 | CA2 | 12332 | 0.060 | 0.3074 | No |
| 121 | ACSM3 | 12384 | 0.059 | 0.3056 | No |
| 122 | FABP1 | 12730 | 0.054 | 0.2873 | No |
| 123 | FABP2 | 12743 | 0.054 | 0.2876 | No |
| 124 | INMT | 13287 | 0.047 | 0.2580 | No |
| 125 | ADIPOR2 | 13387 | 0.045 | 0.2533 | No |
| 126 | FASN | 13470 | 0.044 | 0.2495 | No |
| 127 | SLC22A5 | 13552 | 0.043 | 0.2457 | No |
| 128 | CBR1 | 14017 | 0.036 | 0.2204 | No |
| 129 | EPHX1 | 14047 | 0.035 | 0.2194 | No |
| 130 | GAD2 | 14227 | 0.032 | 0.2099 | No |
| 131 | H2AZ1 | 14333 | 0.027 | 0.2045 | No |
| 132 | HCCS | 14543 | -0.000 | 0.1928 | No |
| 133 | ME1 | 14685 | -0.000 | 0.1849 | No |
| 134 | IDH3G | 14701 | -0.000 | 0.1841 | No |
| 135 | ACSL4 | 14707 | -0.000 | 0.1838 | No |
| 136 | HSP90AA1 | 14786 | -0.000 | 0.1794 | No |
| 137 | ERP29 | 14844 | -0.000 | 0.1763 | No |
| 138 | IDH3B | 14953 | -0.000 | 0.1702 | No |
| 139 | HMGCS1 | 15258 | -0.000 | 0.1532 | No |
| 140 | GPD2 | 15307 | -0.000 | 0.1505 | No |
| 141 | REEP6 | 15312 | -0.000 | 0.1503 | No |
| 142 | YWHAH | 15629 | -0.000 | 0.1326 | No |
| 143 | BMPR1B | 15919 | -0.000 | 0.1164 | No |
| 144 | HADH | 15926 | -0.000 | 0.1160 | No |
| 145 | ALDH1A1 | 16547 | -0.000 | 0.0813 | No |
| 146 | KMT5A | 17215 | -0.000 | 0.0440 | No |
| 147 | MAOA | 17408 | -0.000 | 0.0332 | No |
| 148 | S100A10 | 17513 | -0.000 | 0.0274 | No |
| 149 | SDHD | 17623 | -0.000 | 0.0213 | No |