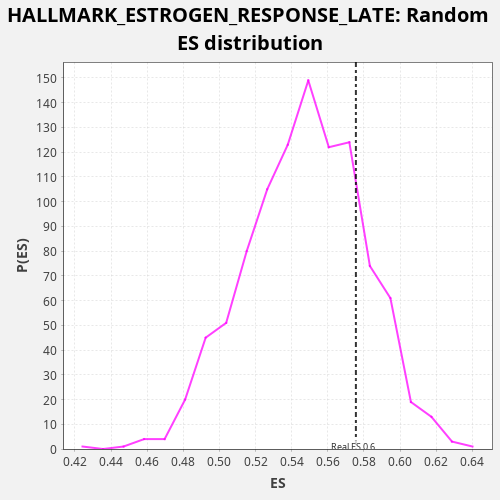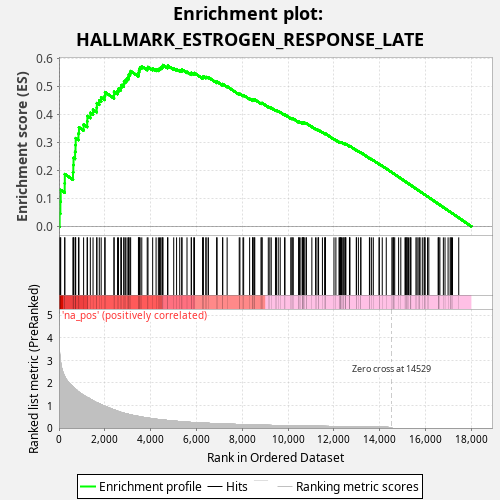
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_ESTROGEN_RESPONSE_LATE |
| Enrichment Score (ES) | 0.57555026 |
| Normalized Enrichment Score (NES) | 1.0512191 |
| Nominal p-value | 0.195 |
| FDR q-value | 0.6107503 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | JAK2 | 29 | 3.319 | 0.0469 | Yes |
| 2 | HSPA4L | 60 | 2.980 | 0.0887 | Yes |
| 3 | FDFT1 | 75 | 2.893 | 0.1302 | Yes |
| 4 | CELSR2 | 249 | 2.286 | 0.1539 | Yes |
| 5 | CHPT1 | 254 | 2.257 | 0.1866 | Yes |
| 6 | PERP | 611 | 1.821 | 0.1932 | Yes |
| 7 | SLC24A3 | 625 | 1.803 | 0.2188 | Yes |
| 8 | ELOVL5 | 635 | 1.791 | 0.2445 | Yes |
| 9 | SORD | 702 | 1.736 | 0.2662 | Yes |
| 10 | IL6ST | 721 | 1.721 | 0.2903 | Yes |
| 11 | MAPT | 729 | 1.712 | 0.3149 | Yes |
| 12 | SERPINA3 | 850 | 1.602 | 0.3316 | Yes |
| 13 | TFF1 | 872 | 1.584 | 0.3535 | Yes |
| 14 | FKBP5 | 1074 | 1.450 | 0.3634 | Yes |
| 15 | IDH2 | 1231 | 1.354 | 0.3745 | Yes |
| 16 | GALE | 1238 | 1.351 | 0.3938 | Yes |
| 17 | CDC6 | 1368 | 1.274 | 0.4052 | Yes |
| 18 | SLC26A2 | 1486 | 1.212 | 0.4164 | Yes |
| 19 | RABEP1 | 1648 | 1.120 | 0.4237 | Yes |
| 20 | AMFR | 1653 | 1.118 | 0.4398 | Yes |
| 21 | RPS6KA2 | 1759 | 1.071 | 0.4495 | Yes |
| 22 | IMPA2 | 1841 | 1.035 | 0.4601 | Yes |
| 23 | ACOX2 | 1998 | 0.964 | 0.4654 | Yes |
| 24 | KLK11 | 2011 | 0.960 | 0.4788 | Yes |
| 25 | IGSF1 | 2401 | 0.806 | 0.4687 | Yes |
| 26 | METTL3 | 2404 | 0.805 | 0.4804 | Yes |
| 27 | ALDH3B1 | 2555 | 0.744 | 0.4828 | Yes |
| 28 | HOMER2 | 2597 | 0.730 | 0.4912 | Yes |
| 29 | ST6GALNAC2 | 2698 | 0.693 | 0.4957 | Yes |
| 30 | MYOF | 2725 | 0.683 | 0.5042 | Yes |
| 31 | BTG3 | 2826 | 0.655 | 0.5082 | Yes |
| 32 | FGFR3 | 2841 | 0.651 | 0.5169 | Yes |
| 33 | PAPSS2 | 2909 | 0.635 | 0.5224 | Yes |
| 34 | TPBG | 2974 | 0.618 | 0.5278 | Yes |
| 35 | LTF | 3036 | 0.603 | 0.5332 | Yes |
| 36 | XRCC3 | 3048 | 0.600 | 0.5414 | Yes |
| 37 | CLIC3 | 3097 | 0.588 | 0.5473 | Yes |
| 38 | MAPK13 | 3128 | 0.580 | 0.5540 | Yes |
| 39 | DHCR7 | 3468 | 0.510 | 0.5425 | Yes |
| 40 | LLGL2 | 3480 | 0.507 | 0.5493 | Yes |
| 41 | CALCR | 3496 | 0.503 | 0.5558 | Yes |
| 42 | FARP1 | 3511 | 0.501 | 0.5623 | Yes |
| 43 | SCUBE2 | 3543 | 0.496 | 0.5678 | Yes |
| 44 | PDZK1 | 3614 | 0.482 | 0.5709 | Yes |
| 45 | DNAJC12 | 3851 | 0.442 | 0.5641 | Yes |
| 46 | MYB | 3885 | 0.436 | 0.5686 | Yes |
| 47 | LSR | 4089 | 0.407 | 0.5632 | Yes |
| 48 | ANXA9 | 4238 | 0.386 | 0.5605 | Yes |
| 49 | CA12 | 4338 | 0.373 | 0.5604 | Yes |
| 50 | SLC2A8 | 4386 | 0.368 | 0.5631 | Yes |
| 51 | PLAAT3 | 4436 | 0.362 | 0.5657 | Yes |
| 52 | STIL | 4494 | 0.356 | 0.5677 | Yes |
| 53 | ALDH3A2 | 4522 | 0.353 | 0.5713 | Yes |
| 54 | TH | 4539 | 0.351 | 0.5756 | Yes |
| 55 | LAMC2 | 4737 | 0.330 | 0.5693 | No |
| 56 | NBL1 | 4754 | 0.329 | 0.5732 | No |
| 57 | PKP3 | 5008 | 0.305 | 0.5635 | No |
| 58 | BAG1 | 5133 | 0.294 | 0.5608 | No |
| 59 | SCARB1 | 5270 | 0.281 | 0.5573 | No |
| 60 | TRIM29 | 5358 | 0.274 | 0.5564 | No |
| 61 | TST | 5370 | 0.273 | 0.5598 | No |
| 62 | TOP2A | 5588 | 0.258 | 0.5514 | No |
| 63 | PDCD4 | 5775 | 0.244 | 0.5445 | No |
| 64 | MOCS2 | 5778 | 0.244 | 0.5479 | No |
| 65 | TFPI2 | 5883 | 0.237 | 0.5456 | No |
| 66 | PPIF | 5916 | 0.236 | 0.5472 | No |
| 67 | TPSAB1 | 6269 | 0.217 | 0.5306 | No |
| 68 | TJP3 | 6286 | 0.216 | 0.5329 | No |
| 69 | PLXNB1 | 6302 | 0.215 | 0.5352 | No |
| 70 | FRK | 6406 | 0.210 | 0.5325 | No |
| 71 | TMPRSS3 | 6455 | 0.208 | 0.5328 | No |
| 72 | PLK4 | 6524 | 0.204 | 0.5320 | No |
| 73 | AGR2 | 6881 | 0.188 | 0.5147 | No |
| 74 | BLVRB | 6901 | 0.187 | 0.5164 | No |
| 75 | UNC13B | 7143 | 0.177 | 0.5054 | No |
| 76 | GPER1 | 7147 | 0.176 | 0.5079 | No |
| 77 | CCN5 | 7340 | 0.169 | 0.4995 | No |
| 78 | RBBP8 | 7868 | 0.152 | 0.4722 | No |
| 79 | GINS2 | 7898 | 0.151 | 0.4728 | No |
| 80 | HR | 8032 | 0.146 | 0.4674 | No |
| 81 | IGFBP4 | 8061 | 0.145 | 0.4680 | No |
| 82 | CHST8 | 8322 | 0.137 | 0.4554 | No |
| 83 | EMP2 | 8451 | 0.133 | 0.4501 | No |
| 84 | UGDH | 8465 | 0.133 | 0.4513 | No |
| 85 | IL17RB | 8471 | 0.133 | 0.4530 | No |
| 86 | CISH | 8527 | 0.131 | 0.4518 | No |
| 87 | PDLIM3 | 8547 | 0.131 | 0.4527 | No |
| 88 | CYP4F11 | 8818 | 0.124 | 0.4393 | No |
| 89 | SULT2B1 | 8855 | 0.123 | 0.4391 | No |
| 90 | OPN3 | 8867 | 0.123 | 0.4403 | No |
| 91 | SOX3 | 9137 | 0.115 | 0.4269 | No |
| 92 | TIAM1 | 9204 | 0.114 | 0.4248 | No |
| 93 | SLC7A5 | 9265 | 0.112 | 0.4231 | No |
| 94 | BATF | 9451 | 0.108 | 0.4143 | No |
| 95 | TPD52L1 | 9493 | 0.107 | 0.4136 | No |
| 96 | KRT13 | 9571 | 0.106 | 0.4108 | No |
| 97 | KRT19 | 9657 | 0.104 | 0.4075 | No |
| 98 | LARGE1 | 9847 | 0.100 | 0.3984 | No |
| 99 | GJB3 | 9866 | 0.099 | 0.3988 | No |
| 100 | DCXR | 10118 | 0.095 | 0.3861 | No |
| 101 | SLC16A1 | 10170 | 0.094 | 0.3846 | No |
| 102 | ST14 | 10178 | 0.094 | 0.3856 | No |
| 103 | KLK10 | 10227 | 0.093 | 0.3842 | No |
| 104 | KCNK5 | 10463 | 0.089 | 0.3723 | No |
| 105 | AREG | 10470 | 0.089 | 0.3733 | No |
| 106 | ARL3 | 10503 | 0.088 | 0.3728 | No |
| 107 | PRSS23 | 10561 | 0.087 | 0.3709 | No |
| 108 | DNAJC1 | 10642 | 0.086 | 0.3676 | No |
| 109 | SLC29A1 | 10644 | 0.086 | 0.3688 | No |
| 110 | XBP1 | 10646 | 0.086 | 0.3700 | No |
| 111 | ADD3 | 10655 | 0.085 | 0.3708 | No |
| 112 | DHRS2 | 10683 | 0.085 | 0.3705 | No |
| 113 | HMGCS2 | 10704 | 0.085 | 0.3707 | No |
| 114 | SERPINA1 | 10787 | 0.083 | 0.3673 | No |
| 115 | FLNB | 10799 | 0.083 | 0.3679 | No |
| 116 | MICB | 11039 | 0.078 | 0.3556 | No |
| 117 | TNNC1 | 11200 | 0.076 | 0.3477 | No |
| 118 | FKBP4 | 11260 | 0.075 | 0.3455 | No |
| 119 | ATP2B4 | 11324 | 0.074 | 0.3431 | No |
| 120 | RNASEH2A | 11332 | 0.074 | 0.3437 | No |
| 121 | DLG5 | 11500 | 0.072 | 0.3354 | No |
| 122 | CAV1 | 11600 | 0.070 | 0.3309 | No |
| 123 | ETFB | 11616 | 0.070 | 0.3311 | No |
| 124 | SGK1 | 11633 | 0.069 | 0.3312 | No |
| 125 | SNX10 | 12004 | 0.064 | 0.3113 | No |
| 126 | NMU | 12088 | 0.063 | 0.3076 | No |
| 127 | SERPINA5 | 12226 | 0.061 | 0.3008 | No |
| 128 | SCNN1A | 12242 | 0.061 | 0.3008 | No |
| 129 | COX6C | 12283 | 0.060 | 0.2995 | No |
| 130 | CACNA2D2 | 12286 | 0.060 | 0.3002 | No |
| 131 | PTPN6 | 12309 | 0.060 | 0.2999 | No |
| 132 | CA2 | 12332 | 0.060 | 0.2995 | No |
| 133 | ISG20 | 12387 | 0.059 | 0.2973 | No |
| 134 | MEST | 12438 | 0.058 | 0.2954 | No |
| 135 | PGR | 12440 | 0.058 | 0.2962 | No |
| 136 | PRLR | 12495 | 0.057 | 0.2940 | No |
| 137 | CDH1 | 12514 | 0.057 | 0.2938 | No |
| 138 | AFF1 | 12520 | 0.057 | 0.2943 | No |
| 139 | DUSP2 | 12681 | 0.055 | 0.2861 | No |
| 140 | RAB31 | 12691 | 0.055 | 0.2864 | No |
| 141 | HSPB8 | 12697 | 0.054 | 0.2870 | No |
| 142 | GAL | 12981 | 0.051 | 0.2718 | No |
| 143 | TFF3 | 12984 | 0.051 | 0.2724 | No |
| 144 | ABHD2 | 13070 | 0.049 | 0.2684 | No |
| 145 | FABP5 | 13175 | 0.048 | 0.2632 | No |
| 146 | NPY1R | 13176 | 0.048 | 0.2639 | No |
| 147 | SLC22A5 | 13552 | 0.043 | 0.2435 | No |
| 148 | S100A9 | 13568 | 0.042 | 0.2433 | No |
| 149 | CPE | 13648 | 0.042 | 0.2395 | No |
| 150 | PRKAR2B | 13722 | 0.041 | 0.2360 | No |
| 151 | JAK1 | 13977 | 0.036 | 0.2222 | No |
| 152 | CKB | 13979 | 0.036 | 0.2227 | No |
| 153 | RAPGEFL1 | 14112 | 0.034 | 0.2158 | No |
| 154 | GLA | 14287 | 0.029 | 0.2064 | No |
| 155 | CYP26B1 | 14533 | -0.000 | 0.1927 | No |
| 156 | CD9 | 14579 | -0.000 | 0.1902 | No |
| 157 | SEMA3B | 14591 | -0.000 | 0.1895 | No |
| 158 | CD44 | 14614 | -0.000 | 0.1883 | No |
| 159 | PTGER3 | 14656 | -0.000 | 0.1860 | No |
| 160 | TFAP2C | 14826 | -0.000 | 0.1765 | No |
| 161 | ITPK1 | 14922 | -0.000 | 0.1712 | No |
| 162 | TSPAN13 | 15105 | -0.000 | 0.1610 | No |
| 163 | CXCL12 | 15128 | -0.000 | 0.1597 | No |
| 164 | WFS1 | 15183 | -0.000 | 0.1567 | No |
| 165 | CCND1 | 15191 | -0.000 | 0.1563 | No |
| 166 | MDK | 15197 | -0.000 | 0.1560 | No |
| 167 | KIF20A | 15259 | -0.000 | 0.1526 | No |
| 168 | ID2 | 15330 | -0.000 | 0.1487 | No |
| 169 | SLC1A4 | 15334 | -0.000 | 0.1485 | No |
| 170 | CDC20 | 15366 | -0.000 | 0.1468 | No |
| 171 | OVOL2 | 15583 | -0.000 | 0.1346 | No |
| 172 | ZFP36 | 15627 | -0.000 | 0.1322 | No |
| 173 | OLFM1 | 15679 | -0.000 | 0.1294 | No |
| 174 | NXT1 | 15748 | -0.000 | 0.1255 | No |
| 175 | CCNA1 | 15758 | -0.000 | 0.1250 | No |
| 176 | KLF4 | 15860 | -0.000 | 0.1194 | No |
| 177 | ASCL1 | 15938 | -0.000 | 0.1150 | No |
| 178 | SLC27A2 | 15965 | -0.000 | 0.1136 | No |
| 179 | TOB1 | 15983 | -0.000 | 0.1126 | No |
| 180 | CXCL14 | 16089 | -0.000 | 0.1067 | No |
| 181 | PTGES | 16145 | -0.000 | 0.1036 | No |
| 182 | DYNLT3 | 16551 | -0.000 | 0.0809 | No |
| 183 | HPRT1 | 16577 | -0.000 | 0.0795 | No |
| 184 | RET | 16579 | -0.000 | 0.0794 | No |
| 185 | NAB2 | 16628 | -0.000 | 0.0768 | No |
| 186 | FOS | 16780 | -0.000 | 0.0683 | No |
| 187 | BCL2 | 16852 | -0.000 | 0.0643 | No |
| 188 | SFN | 16986 | -0.000 | 0.0568 | No |
| 189 | EGR3 | 17076 | -0.000 | 0.0518 | No |
| 190 | NRIP1 | 17108 | -0.000 | 0.0501 | No |
| 191 | SIAH2 | 17137 | -0.000 | 0.0485 | No |
| 192 | PCP4 | 17179 | -0.000 | 0.0462 | No |
| 193 | NCOR2 | 17451 | -0.000 | 0.0310 | No |