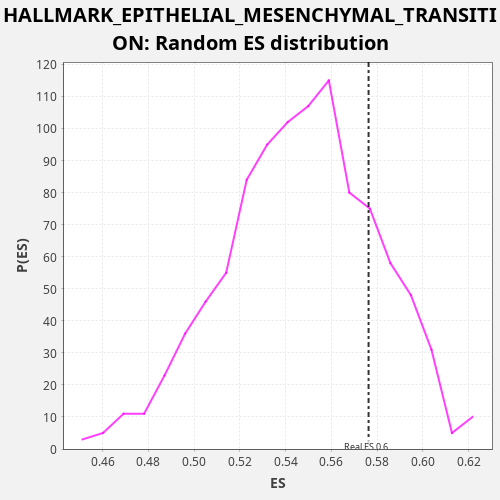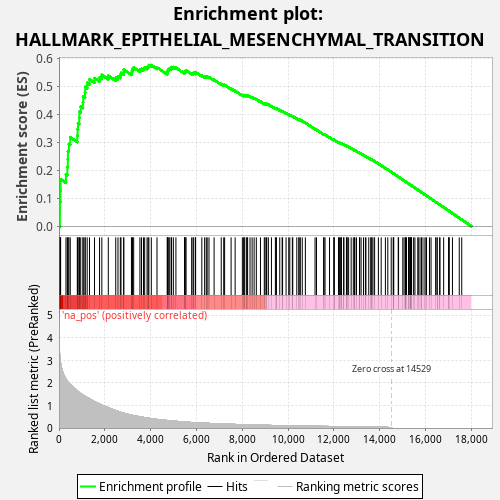
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | 0.5762298 |
| Normalized Enrichment Score (NES) | 1.0540587 |
| Nominal p-value | 0.192 |
| FDR q-value | 0.6086317 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | FERMT2 | 40 | 3.191 | 0.0428 | Yes |
| 2 | MXRA5 | 41 | 3.163 | 0.0875 | Yes |
| 3 | TNFRSF11B | 63 | 2.939 | 0.1278 | Yes |
| 4 | COL6A2 | 71 | 2.902 | 0.1684 | Yes |
| 5 | LRP1 | 304 | 2.171 | 0.1860 | Yes |
| 6 | CDH11 | 364 | 2.082 | 0.2121 | Yes |
| 7 | MMP14 | 384 | 2.049 | 0.2400 | Yes |
| 8 | CALD1 | 390 | 2.039 | 0.2685 | Yes |
| 9 | VIM | 433 | 1.995 | 0.2943 | Yes |
| 10 | COL11A1 | 491 | 1.928 | 0.3183 | Yes |
| 11 | BGN | 799 | 1.643 | 0.3243 | Yes |
| 12 | SFRP4 | 812 | 1.631 | 0.3466 | Yes |
| 13 | COL8A2 | 831 | 1.617 | 0.3684 | Yes |
| 14 | TIMP3 | 884 | 1.573 | 0.3877 | Yes |
| 15 | THY1 | 891 | 1.571 | 0.4096 | Yes |
| 16 | DST | 948 | 1.531 | 0.4281 | Yes |
| 17 | SLIT3 | 1046 | 1.465 | 0.4433 | Yes |
| 18 | PRSS2 | 1056 | 1.461 | 0.4634 | Yes |
| 19 | LAMA1 | 1135 | 1.406 | 0.4789 | Yes |
| 20 | COL16A1 | 1144 | 1.401 | 0.4982 | Yes |
| 21 | GAS1 | 1230 | 1.354 | 0.5126 | Yes |
| 22 | SDC1 | 1332 | 1.302 | 0.5253 | Yes |
| 23 | GLIPR1 | 1550 | 1.169 | 0.5296 | Yes |
| 24 | SLIT2 | 1770 | 1.068 | 0.5324 | Yes |
| 25 | PTX3 | 1870 | 1.019 | 0.5412 | Yes |
| 26 | ANPEP | 2152 | 0.903 | 0.5382 | Yes |
| 27 | SPP1 | 2477 | 0.773 | 0.5309 | Yes |
| 28 | FUCA1 | 2575 | 0.736 | 0.5359 | Yes |
| 29 | ELN | 2677 | 0.700 | 0.5401 | Yes |
| 30 | CALU | 2707 | 0.689 | 0.5482 | Yes |
| 31 | THBS2 | 2825 | 0.655 | 0.5509 | Yes |
| 32 | LOXL2 | 2827 | 0.654 | 0.5600 | Yes |
| 33 | ITGB3 | 3163 | 0.573 | 0.5493 | Yes |
| 34 | GEM | 3188 | 0.569 | 0.5560 | Yes |
| 35 | BMP1 | 3211 | 0.560 | 0.5627 | Yes |
| 36 | COL6A3 | 3260 | 0.549 | 0.5677 | Yes |
| 37 | COL5A3 | 3533 | 0.497 | 0.5595 | Yes |
| 38 | COL12A1 | 3602 | 0.484 | 0.5625 | Yes |
| 39 | ECM1 | 3688 | 0.467 | 0.5643 | Yes |
| 40 | FGF2 | 3740 | 0.458 | 0.5679 | Yes |
| 41 | OXTR | 3845 | 0.443 | 0.5684 | Yes |
| 42 | SGCG | 3892 | 0.434 | 0.5719 | Yes |
| 43 | TPM2 | 3924 | 0.430 | 0.5762 | Yes |
| 44 | FBLN2 | 4030 | 0.417 | 0.5762 | No |
| 45 | MGP | 4278 | 0.381 | 0.5677 | No |
| 46 | IL32 | 4723 | 0.332 | 0.5475 | No |
| 47 | LAMC2 | 4737 | 0.330 | 0.5514 | No |
| 48 | HTRA1 | 4745 | 0.330 | 0.5557 | No |
| 49 | MYLK | 4760 | 0.329 | 0.5595 | No |
| 50 | CAPG | 4799 | 0.325 | 0.5620 | No |
| 51 | PCOLCE2 | 4839 | 0.321 | 0.5643 | No |
| 52 | ECM2 | 4907 | 0.314 | 0.5650 | No |
| 53 | QSOX1 | 4914 | 0.314 | 0.5691 | No |
| 54 | VEGFA | 4992 | 0.306 | 0.5691 | No |
| 55 | RGS4 | 5104 | 0.297 | 0.5671 | No |
| 56 | FN1 | 5479 | 0.265 | 0.5498 | No |
| 57 | CXCL8 | 5482 | 0.265 | 0.5534 | No |
| 58 | PLOD2 | 5533 | 0.261 | 0.5543 | No |
| 59 | GPX7 | 5542 | 0.261 | 0.5575 | No |
| 60 | GADD45B | 5791 | 0.243 | 0.5470 | No |
| 61 | MATN3 | 5850 | 0.240 | 0.5472 | No |
| 62 | TFPI2 | 5883 | 0.237 | 0.5487 | No |
| 63 | COLGALT1 | 5961 | 0.233 | 0.5477 | No |
| 64 | PCOLCE | 5963 | 0.233 | 0.5509 | No |
| 65 | PLOD1 | 6235 | 0.219 | 0.5388 | No |
| 66 | SNTB1 | 6358 | 0.212 | 0.5349 | No |
| 67 | PVR | 6428 | 0.209 | 0.5340 | No |
| 68 | MMP3 | 6468 | 0.207 | 0.5348 | No |
| 69 | MFAP5 | 6547 | 0.203 | 0.5332 | No |
| 70 | COMP | 6771 | 0.192 | 0.5234 | No |
| 71 | LAMA2 | 7082 | 0.179 | 0.5086 | No |
| 72 | P3H1 | 7191 | 0.175 | 0.5050 | No |
| 73 | MATN2 | 7230 | 0.173 | 0.5053 | No |
| 74 | CXCL6 | 7513 | 0.163 | 0.4917 | No |
| 75 | CRLF1 | 7689 | 0.157 | 0.4841 | No |
| 76 | COL4A2 | 8004 | 0.147 | 0.4686 | No |
| 77 | IGFBP4 | 8061 | 0.145 | 0.4675 | No |
| 78 | BDNF | 8078 | 0.145 | 0.4686 | No |
| 79 | PDGFRB | 8110 | 0.144 | 0.4689 | No |
| 80 | FBN2 | 8196 | 0.141 | 0.4661 | No |
| 81 | TGFBR3 | 8205 | 0.141 | 0.4677 | No |
| 82 | ITGB5 | 8234 | 0.139 | 0.4681 | No |
| 83 | CTHRC1 | 8339 | 0.136 | 0.4642 | No |
| 84 | FBLN1 | 8430 | 0.134 | 0.4610 | No |
| 85 | CAP2 | 8518 | 0.132 | 0.4580 | No |
| 86 | SDC4 | 8614 | 0.129 | 0.4544 | No |
| 87 | LAMA3 | 8796 | 0.124 | 0.4460 | No |
| 88 | NT5E | 8962 | 0.120 | 0.4385 | No |
| 89 | APLP1 | 8998 | 0.119 | 0.4382 | No |
| 90 | PPIB | 9055 | 0.118 | 0.4367 | No |
| 91 | SGCB | 9069 | 0.117 | 0.4376 | No |
| 92 | TNFRSF12A | 9140 | 0.115 | 0.4353 | No |
| 93 | FZD8 | 9274 | 0.112 | 0.4294 | No |
| 94 | LOXL1 | 9446 | 0.108 | 0.4214 | No |
| 95 | IGFBP2 | 9481 | 0.108 | 0.4210 | No |
| 96 | SGCD | 9490 | 0.107 | 0.4220 | No |
| 97 | COL7A1 | 9637 | 0.104 | 0.4153 | No |
| 98 | TNC | 9741 | 0.102 | 0.4110 | No |
| 99 | SERPINH1 | 9761 | 0.101 | 0.4113 | No |
| 100 | MCM7 | 9913 | 0.099 | 0.4043 | No |
| 101 | MMP1 | 10029 | 0.096 | 0.3992 | No |
| 102 | ITGA5 | 10076 | 0.096 | 0.3979 | No |
| 103 | LGALS1 | 10187 | 0.094 | 0.3931 | No |
| 104 | EMP3 | 10206 | 0.093 | 0.3934 | No |
| 105 | VCAM1 | 10381 | 0.090 | 0.3849 | No |
| 106 | AREG | 10470 | 0.089 | 0.3812 | No |
| 107 | SLC6A8 | 10483 | 0.089 | 0.3818 | No |
| 108 | TGFBI | 10535 | 0.088 | 0.3801 | No |
| 109 | MMP2 | 10620 | 0.086 | 0.3766 | No |
| 110 | COL1A1 | 10755 | 0.084 | 0.3703 | No |
| 111 | THBS1 | 11178 | 0.077 | 0.3477 | No |
| 112 | ITGAV | 11241 | 0.076 | 0.3453 | No |
| 113 | EFEMP2 | 11551 | 0.071 | 0.3289 | No |
| 114 | COL4A1 | 11569 | 0.071 | 0.3290 | No |
| 115 | LRRC15 | 11614 | 0.070 | 0.3275 | No |
| 116 | FAP | 11810 | 0.067 | 0.3175 | No |
| 117 | NOTCH2 | 12003 | 0.064 | 0.3076 | No |
| 118 | PLAUR | 12009 | 0.064 | 0.3082 | No |
| 119 | ENO2 | 12012 | 0.064 | 0.3090 | No |
| 120 | VCAN | 12201 | 0.061 | 0.2993 | No |
| 121 | COL1A2 | 12228 | 0.061 | 0.2987 | No |
| 122 | DAB2 | 12238 | 0.061 | 0.2990 | No |
| 123 | FSTL3 | 12296 | 0.060 | 0.2967 | No |
| 124 | GPC1 | 12324 | 0.060 | 0.2960 | No |
| 125 | ADAM12 | 12331 | 0.060 | 0.2965 | No |
| 126 | SCG2 | 12425 | 0.058 | 0.2921 | No |
| 127 | MEST | 12438 | 0.058 | 0.2923 | No |
| 128 | TAGLN | 12548 | 0.056 | 0.2869 | No |
| 129 | FSTL1 | 12566 | 0.056 | 0.2868 | No |
| 130 | MYL9 | 12581 | 0.056 | 0.2868 | No |
| 131 | LAMC1 | 12644 | 0.055 | 0.2841 | No |
| 132 | TGM2 | 12754 | 0.054 | 0.2787 | No |
| 133 | TPM4 | 12848 | 0.052 | 0.2742 | No |
| 134 | DPYSL3 | 12904 | 0.052 | 0.2719 | No |
| 135 | IGFBP3 | 12916 | 0.051 | 0.2720 | No |
| 136 | FBLN5 | 12987 | 0.051 | 0.2688 | No |
| 137 | COL5A2 | 13119 | 0.049 | 0.2621 | No |
| 138 | ITGB1 | 13126 | 0.049 | 0.2624 | No |
| 139 | TPM1 | 13188 | 0.048 | 0.2597 | No |
| 140 | CXCL1 | 13293 | 0.047 | 0.2545 | No |
| 141 | WIPF1 | 13376 | 0.046 | 0.2505 | No |
| 142 | COL3A1 | 13400 | 0.045 | 0.2499 | No |
| 143 | GJA1 | 13523 | 0.043 | 0.2437 | No |
| 144 | PDLIM4 | 13613 | 0.042 | 0.2392 | No |
| 145 | CDH6 | 13624 | 0.042 | 0.2393 | No |
| 146 | SPOCK1 | 13660 | 0.041 | 0.2379 | No |
| 147 | FBN1 | 13706 | 0.041 | 0.2359 | No |
| 148 | IL15 | 13778 | 0.040 | 0.2325 | No |
| 149 | FMOD | 13943 | 0.037 | 0.2238 | No |
| 150 | SFRP1 | 14065 | 0.035 | 0.2175 | No |
| 151 | NTM | 14249 | 0.031 | 0.2077 | No |
| 152 | INHBA | 14354 | 0.026 | 0.2022 | No |
| 153 | PFN2 | 14503 | 0.015 | 0.1941 | No |
| 154 | DCN | 14589 | -0.000 | 0.1894 | No |
| 155 | SNAI2 | 14604 | -0.000 | 0.1886 | No |
| 156 | CD44 | 14614 | -0.000 | 0.1881 | No |
| 157 | CD59 | 14809 | -0.000 | 0.1772 | No |
| 158 | PTHLH | 14824 | -0.000 | 0.1764 | No |
| 159 | TIMP1 | 15009 | -0.000 | 0.1660 | No |
| 160 | TGFB1 | 15066 | -0.000 | 0.1629 | No |
| 161 | CXCL12 | 15128 | -0.000 | 0.1595 | No |
| 162 | ACTA2 | 15133 | -0.000 | 0.1593 | No |
| 163 | DKK1 | 15140 | -0.000 | 0.1589 | No |
| 164 | PMP22 | 15171 | -0.000 | 0.1572 | No |
| 165 | LOX | 15264 | -0.000 | 0.1521 | No |
| 166 | SPARC | 15265 | -0.000 | 0.1521 | No |
| 167 | WNT5A | 15291 | -0.000 | 0.1507 | No |
| 168 | ID2 | 15330 | -0.000 | 0.1485 | No |
| 169 | PRRX1 | 15338 | -0.000 | 0.1481 | No |
| 170 | GADD45A | 15351 | -0.000 | 0.1475 | No |
| 171 | TNFAIP3 | 15390 | -0.000 | 0.1453 | No |
| 172 | CCN2 | 15391 | -0.000 | 0.1453 | No |
| 173 | COPA | 15481 | -0.000 | 0.1403 | No |
| 174 | PMEPA1 | 15538 | -0.000 | 0.1372 | No |
| 175 | SAT1 | 15668 | -0.000 | 0.1299 | No |
| 176 | COL5A1 | 15681 | -0.000 | 0.1293 | No |
| 177 | POSTN | 15760 | -0.000 | 0.1249 | No |
| 178 | SERPINE2 | 15830 | -0.000 | 0.1210 | No |
| 179 | IL6 | 15843 | -0.000 | 0.1203 | No |
| 180 | LUM | 15937 | -0.000 | 0.1151 | No |
| 181 | CCN1 | 16014 | -0.000 | 0.1108 | No |
| 182 | RHOB | 16045 | -0.000 | 0.1092 | No |
| 183 | VEGFC | 16174 | -0.000 | 0.1020 | No |
| 184 | ABI3BP | 16242 | -0.000 | 0.0982 | No |
| 185 | MSX1 | 16442 | -0.000 | 0.0870 | No |
| 186 | ITGA2 | 16506 | -0.000 | 0.0835 | No |
| 187 | EDIL3 | 16507 | -0.000 | 0.0835 | No |
| 188 | NNMT | 16618 | -0.000 | 0.0773 | No |
| 189 | GREM1 | 16631 | -0.000 | 0.0767 | No |
| 190 | CDH2 | 16792 | -0.000 | 0.0677 | No |
| 191 | FOXC2 | 17009 | -0.000 | 0.0555 | No |
| 192 | BASP1 | 17014 | -0.000 | 0.0553 | No |
| 193 | JUN | 17035 | -0.000 | 0.0542 | No |
| 194 | CADM1 | 17176 | -0.000 | 0.0463 | No |
| 195 | FLNA | 17474 | -0.000 | 0.0296 | No |
| 196 | MAGEE1 | 17582 | -0.000 | 0.0236 | No |