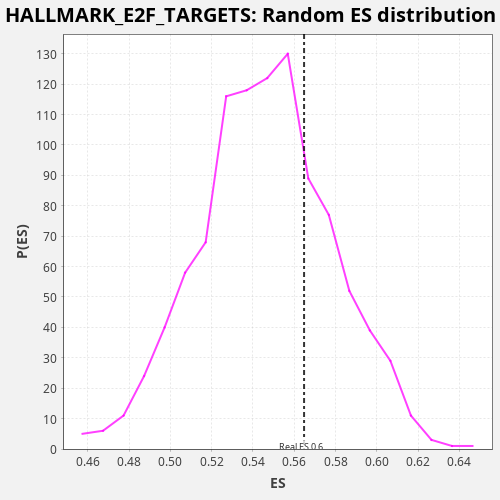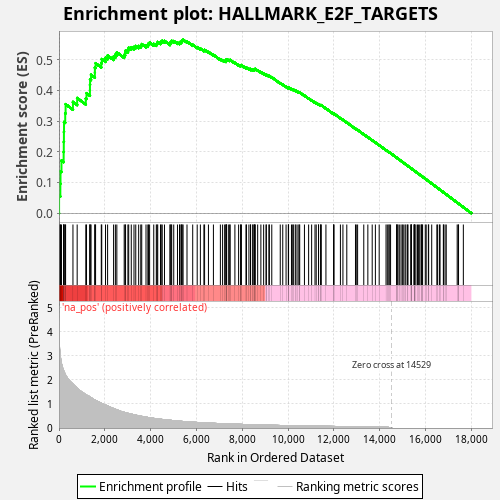
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_E2F_TARGETS |
| Enrichment Score (ES) | 0.5647472 |
| Normalized Enrichment Score (NES) | 1.0339656 |
| Nominal p-value | 0.279 |
| FDR q-value | 0.71304363 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | MKI67 | 4 | 4.130 | 0.0572 | Yes |
| 2 | DONSON | 59 | 3.013 | 0.0961 | Yes |
| 3 | MMS22L | 66 | 2.928 | 0.1365 | Yes |
| 4 | CBX5 | 108 | 2.688 | 0.1716 | Yes |
| 5 | CNOT9 | 203 | 2.385 | 0.1995 | Yes |
| 6 | CCNE1 | 209 | 2.370 | 0.2322 | Yes |
| 7 | ING3 | 215 | 2.355 | 0.2647 | Yes |
| 8 | RFC1 | 220 | 2.338 | 0.2970 | Yes |
| 9 | TIPIN | 272 | 2.229 | 0.3251 | Yes |
| 10 | MSH2 | 288 | 2.197 | 0.3549 | Yes |
| 11 | LIG1 | 609 | 1.822 | 0.3622 | Yes |
| 12 | SMC3 | 793 | 1.654 | 0.3750 | Yes |
| 13 | POP7 | 1170 | 1.390 | 0.3732 | Yes |
| 14 | MRE11 | 1201 | 1.368 | 0.3905 | Yes |
| 15 | TIMELESS | 1350 | 1.286 | 0.4001 | Yes |
| 16 | CENPE | 1352 | 1.284 | 0.4179 | Yes |
| 17 | MXD3 | 1357 | 1.281 | 0.4355 | Yes |
| 18 | TK1 | 1399 | 1.259 | 0.4508 | Yes |
| 19 | RACGAP1 | 1565 | 1.163 | 0.4577 | Yes |
| 20 | PDS5B | 1567 | 1.162 | 0.4738 | Yes |
| 21 | CENPM | 1597 | 1.147 | 0.4881 | Yes |
| 22 | NME1 | 1846 | 1.031 | 0.4885 | Yes |
| 23 | HUS1 | 1866 | 1.020 | 0.5017 | Yes |
| 24 | MTHFD2 | 2031 | 0.951 | 0.5057 | Yes |
| 25 | NBN | 2122 | 0.915 | 0.5134 | Yes |
| 26 | RAD1 | 2383 | 0.810 | 0.5100 | Yes |
| 27 | CHEK2 | 2461 | 0.779 | 0.5166 | Yes |
| 28 | ZW10 | 2525 | 0.754 | 0.5235 | Yes |
| 29 | GINS4 | 2846 | 0.650 | 0.5146 | Yes |
| 30 | SMC4 | 2886 | 0.640 | 0.5213 | Yes |
| 31 | CHEK1 | 2905 | 0.636 | 0.5291 | Yes |
| 32 | ATAD2 | 3009 | 0.609 | 0.5318 | Yes |
| 33 | BRCA1 | 3043 | 0.601 | 0.5383 | Yes |
| 34 | MCM3 | 3155 | 0.574 | 0.5401 | Yes |
| 35 | BUB1B | 3274 | 0.547 | 0.5411 | Yes |
| 36 | DSCC1 | 3351 | 0.533 | 0.5442 | Yes |
| 37 | MCM4 | 3478 | 0.507 | 0.5442 | Yes |
| 38 | GINS3 | 3570 | 0.490 | 0.5459 | Yes |
| 39 | RAD51AP1 | 3612 | 0.482 | 0.5503 | Yes |
| 40 | ORC6 | 3798 | 0.450 | 0.5462 | Yes |
| 41 | NAA38 | 3880 | 0.437 | 0.5477 | Yes |
| 42 | SPC25 | 3905 | 0.432 | 0.5524 | Yes |
| 43 | AURKA | 3956 | 0.426 | 0.5555 | Yes |
| 44 | SMC6 | 4136 | 0.399 | 0.5510 | Yes |
| 45 | E2F8 | 4248 | 0.385 | 0.5501 | Yes |
| 46 | CCP110 | 4280 | 0.380 | 0.5537 | Yes |
| 47 | SPAG5 | 4310 | 0.376 | 0.5573 | Yes |
| 48 | PMS2 | 4431 | 0.363 | 0.5556 | Yes |
| 49 | BRCA2 | 4461 | 0.360 | 0.5590 | Yes |
| 50 | TACC3 | 4506 | 0.354 | 0.5614 | Yes |
| 51 | NUP107 | 4605 | 0.345 | 0.5607 | Yes |
| 52 | TMPO | 4846 | 0.320 | 0.5517 | Yes |
| 53 | RFC2 | 4855 | 0.319 | 0.5557 | Yes |
| 54 | PAICS | 4897 | 0.315 | 0.5578 | Yes |
| 55 | SHMT1 | 4920 | 0.313 | 0.5609 | Yes |
| 56 | NUP205 | 5009 | 0.305 | 0.5602 | Yes |
| 57 | DEPDC1 | 5172 | 0.291 | 0.5551 | Yes |
| 58 | AURKB | 5261 | 0.282 | 0.5541 | Yes |
| 59 | WDR90 | 5283 | 0.281 | 0.5569 | Yes |
| 60 | POLE4 | 5340 | 0.276 | 0.5575 | Yes |
| 61 | PRIM2 | 5354 | 0.274 | 0.5606 | Yes |
| 62 | DUT | 5379 | 0.272 | 0.5631 | Yes |
| 63 | PSMC3IP | 5417 | 0.270 | 0.5647 | Yes |
| 64 | TOP2A | 5588 | 0.258 | 0.5588 | No |
| 65 | KIF22 | 5837 | 0.240 | 0.5482 | No |
| 66 | LYAR | 6033 | 0.229 | 0.5405 | No |
| 67 | RAD51C | 6172 | 0.222 | 0.5358 | No |
| 68 | DLGAP5 | 6326 | 0.214 | 0.5302 | No |
| 69 | CDCA8 | 6356 | 0.213 | 0.5315 | No |
| 70 | PLK4 | 6524 | 0.204 | 0.5250 | No |
| 71 | CCNB2 | 6741 | 0.193 | 0.5155 | No |
| 72 | TBRG4 | 7045 | 0.181 | 0.5010 | No |
| 73 | CIT | 7142 | 0.177 | 0.4981 | No |
| 74 | NOP56 | 7232 | 0.173 | 0.4955 | No |
| 75 | POLA2 | 7290 | 0.171 | 0.4947 | No |
| 76 | KIF18B | 7293 | 0.171 | 0.4970 | No |
| 77 | PAN2 | 7298 | 0.171 | 0.4991 | No |
| 78 | UNG | 7310 | 0.171 | 0.5009 | No |
| 79 | PRKDC | 7392 | 0.167 | 0.4987 | No |
| 80 | BIRC5 | 7452 | 0.165 | 0.4977 | No |
| 81 | MCM2 | 7471 | 0.165 | 0.4989 | No |
| 82 | AK2 | 7679 | 0.158 | 0.4895 | No |
| 83 | RAD50 | 7841 | 0.153 | 0.4826 | No |
| 84 | POLD1 | 7929 | 0.150 | 0.4798 | No |
| 85 | KIF2C | 7937 | 0.149 | 0.4815 | No |
| 86 | MCM5 | 7963 | 0.149 | 0.4822 | No |
| 87 | RPA1 | 8163 | 0.142 | 0.4730 | No |
| 88 | JPT1 | 8184 | 0.141 | 0.4738 | No |
| 89 | SLBP | 8291 | 0.138 | 0.4698 | No |
| 90 | POLE | 8361 | 0.136 | 0.4678 | No |
| 91 | PSIP1 | 8364 | 0.135 | 0.4695 | No |
| 92 | DDX39A | 8457 | 0.133 | 0.4662 | No |
| 93 | NOLC1 | 8470 | 0.133 | 0.4674 | No |
| 94 | NCAPD2 | 8521 | 0.131 | 0.4664 | No |
| 95 | ESPL1 | 8535 | 0.131 | 0.4675 | No |
| 96 | DEK | 8556 | 0.130 | 0.4682 | No |
| 97 | BARD1 | 8562 | 0.130 | 0.4697 | No |
| 98 | CDKN3 | 8673 | 0.127 | 0.4653 | No |
| 99 | DCTPP1 | 8813 | 0.124 | 0.4593 | No |
| 100 | NUP153 | 8934 | 0.121 | 0.4542 | No |
| 101 | CDC25B | 9039 | 0.118 | 0.4500 | No |
| 102 | ASF1B | 9052 | 0.118 | 0.4510 | No |
| 103 | CDCA3 | 9164 | 0.115 | 0.4463 | No |
| 104 | CDKN1A | 9183 | 0.114 | 0.4469 | No |
| 105 | DIAPH3 | 9289 | 0.112 | 0.4426 | No |
| 106 | DCLRE1B | 9659 | 0.104 | 0.4233 | No |
| 107 | GINS1 | 9768 | 0.101 | 0.4186 | No |
| 108 | MCM7 | 9913 | 0.099 | 0.4119 | No |
| 109 | MELK | 10012 | 0.097 | 0.4078 | No |
| 110 | RPA3 | 10018 | 0.097 | 0.4088 | No |
| 111 | UBR7 | 10022 | 0.097 | 0.4100 | No |
| 112 | TFRC | 10149 | 0.094 | 0.4043 | No |
| 113 | KPNA2 | 10205 | 0.093 | 0.4025 | No |
| 114 | MCM6 | 10229 | 0.093 | 0.4025 | No |
| 115 | PNN | 10313 | 0.091 | 0.3991 | No |
| 116 | RANBP1 | 10347 | 0.091 | 0.3985 | No |
| 117 | PTTG1 | 10431 | 0.090 | 0.3951 | No |
| 118 | POLD3 | 10481 | 0.089 | 0.3935 | No |
| 119 | PPM1D | 10508 | 0.088 | 0.3933 | No |
| 120 | TUBG1 | 10715 | 0.084 | 0.3829 | No |
| 121 | RPA2 | 10898 | 0.081 | 0.3738 | No |
| 122 | LUC7L3 | 11029 | 0.079 | 0.3676 | No |
| 123 | SPC24 | 11176 | 0.077 | 0.3605 | No |
| 124 | MYBL2 | 11229 | 0.076 | 0.3586 | No |
| 125 | PLK1 | 11329 | 0.074 | 0.3541 | No |
| 126 | RNASEH2A | 11332 | 0.074 | 0.3550 | No |
| 127 | TCF19 | 11424 | 0.073 | 0.3509 | No |
| 128 | NASP | 11425 | 0.073 | 0.3519 | No |
| 129 | SSRP1 | 11447 | 0.072 | 0.3518 | No |
| 130 | LBR | 11653 | 0.069 | 0.3412 | No |
| 131 | POLD2 | 11986 | 0.064 | 0.3235 | No |
| 132 | LMNB1 | 11987 | 0.064 | 0.3244 | No |
| 133 | HELLS | 12007 | 0.064 | 0.3242 | No |
| 134 | RFC3 | 12285 | 0.060 | 0.3095 | No |
| 135 | NAP1L1 | 12395 | 0.059 | 0.3041 | No |
| 136 | MAD2L1 | 12564 | 0.056 | 0.2955 | No |
| 137 | ORC2 | 12943 | 0.051 | 0.2750 | No |
| 138 | HMMR | 12976 | 0.051 | 0.2739 | No |
| 139 | RRM2 | 13036 | 0.050 | 0.2713 | No |
| 140 | USP1 | 13305 | 0.046 | 0.2569 | No |
| 141 | GSPT1 | 13483 | 0.044 | 0.2475 | No |
| 142 | TRIP13 | 13673 | 0.041 | 0.2375 | No |
| 143 | UBE2T | 13812 | 0.039 | 0.2303 | No |
| 144 | CDC25A | 13978 | 0.036 | 0.2216 | No |
| 145 | EIF2S1 | 14277 | 0.029 | 0.2052 | No |
| 146 | H2AZ1 | 14333 | 0.027 | 0.2025 | No |
| 147 | SUV39H1 | 14402 | 0.022 | 0.1990 | No |
| 148 | HNRNPD | 14414 | 0.021 | 0.1987 | No |
| 149 | RAN | 14475 | 0.018 | 0.1956 | No |
| 150 | SMC1A | 14739 | -0.000 | 0.1808 | No |
| 151 | EED | 14749 | -0.000 | 0.1803 | No |
| 152 | MLH1 | 14762 | -0.000 | 0.1796 | No |
| 153 | XPO1 | 14798 | -0.000 | 0.1777 | No |
| 154 | KIF4A | 14856 | -0.000 | 0.1745 | No |
| 155 | BRMS1L | 14931 | -0.000 | 0.1703 | No |
| 156 | RBBP7 | 14989 | -0.000 | 0.1671 | No |
| 157 | CTCF | 15028 | -0.000 | 0.1650 | No |
| 158 | EZH2 | 15104 | -0.000 | 0.1608 | No |
| 159 | UBE2S | 15144 | -0.000 | 0.1586 | No |
| 160 | CDKN1B | 15217 | -0.000 | 0.1545 | No |
| 161 | ASF1A | 15234 | -0.000 | 0.1536 | No |
| 162 | CDC20 | 15366 | -0.000 | 0.1463 | No |
| 163 | STMN1 | 15375 | -0.000 | 0.1458 | No |
| 164 | PPP1R8 | 15377 | -0.000 | 0.1458 | No |
| 165 | STAG1 | 15378 | -0.000 | 0.1458 | No |
| 166 | CDKN2C | 15501 | -0.000 | 0.1389 | No |
| 167 | PRDX4 | 15504 | -0.000 | 0.1388 | No |
| 168 | CKS2 | 15522 | -0.000 | 0.1378 | No |
| 169 | CSE1L | 15532 | -0.000 | 0.1373 | No |
| 170 | SNRPB | 15582 | -0.000 | 0.1346 | No |
| 171 | ILF3 | 15652 | -0.000 | 0.1307 | No |
| 172 | DNMT1 | 15690 | -0.000 | 0.1286 | No |
| 173 | PCNA | 15747 | -0.000 | 0.1255 | No |
| 174 | SYNCRIP | 15813 | -0.000 | 0.1218 | No |
| 175 | CDK4 | 15820 | -0.000 | 0.1215 | No |
| 176 | TRA2B | 15854 | -0.000 | 0.1197 | No |
| 177 | MYC | 15873 | -0.000 | 0.1186 | No |
| 178 | TP53 | 15990 | -0.000 | 0.1121 | No |
| 179 | ANP32E | 16030 | -0.000 | 0.1099 | No |
| 180 | PRPS1 | 16125 | -0.000 | 0.1047 | No |
| 181 | CDKN2A | 16138 | -0.000 | 0.1040 | No |
| 182 | DCK | 16271 | -0.000 | 0.0966 | No |
| 183 | HMGB2 | 16496 | -0.000 | 0.0840 | No |
| 184 | RAD21 | 16529 | -0.000 | 0.0822 | No |
| 185 | WEE1 | 16608 | -0.000 | 0.0778 | No |
| 186 | NUDT21 | 16635 | -0.000 | 0.0764 | No |
| 187 | CDK1 | 16779 | -0.000 | 0.0683 | No |
| 188 | PA2G4 | 16789 | -0.000 | 0.0678 | No |
| 189 | CTPS1 | 16853 | -0.000 | 0.0643 | No |
| 190 | CKS1B | 16905 | -0.000 | 0.0614 | No |
| 191 | H2AX | 17383 | -0.000 | 0.0346 | No |
| 192 | TUBB | 17430 | -0.000 | 0.0321 | No |
| 193 | XRCC6 | 17442 | -0.000 | 0.0314 | No |
| 194 | IPO7 | 17652 | -0.000 | 0.0197 | No |