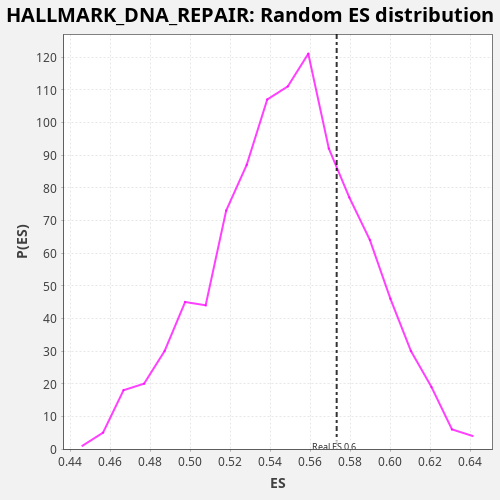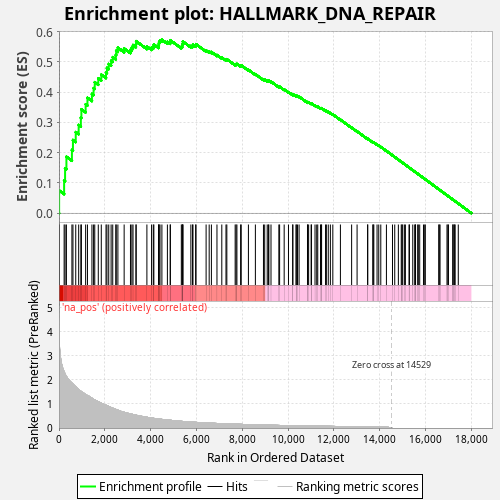
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_DNA_REPAIR |
| Enrichment Score (ES) | 0.5732459 |
| Normalized Enrichment Score (NES) | 1.0446069 |
| Nominal p-value | 0.255 |
| FDR q-value | 0.6252726 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | TSG101 | 6 | 3.922 | 0.0752 | Yes |
| 2 | GTF2B | 224 | 2.327 | 0.1078 | Yes |
| 3 | POLR2H | 266 | 2.237 | 0.1486 | Yes |
| 4 | POLB | 324 | 2.144 | 0.1866 | Yes |
| 5 | IMPDH2 | 564 | 1.867 | 0.2092 | Yes |
| 6 | LIG1 | 609 | 1.822 | 0.2418 | Yes |
| 7 | CSTF3 | 731 | 1.709 | 0.2679 | Yes |
| 8 | ERCC4 | 859 | 1.594 | 0.2915 | Yes |
| 9 | RRM2B | 954 | 1.527 | 0.3156 | Yes |
| 10 | AGO4 | 978 | 1.505 | 0.3433 | Yes |
| 11 | CMPK2 | 1163 | 1.392 | 0.3598 | Yes |
| 12 | CANT1 | 1242 | 1.348 | 0.3814 | Yes |
| 13 | NPR2 | 1438 | 1.236 | 0.3942 | Yes |
| 14 | GTF2H3 | 1510 | 1.190 | 0.4132 | Yes |
| 15 | BCAM | 1561 | 1.164 | 0.4328 | Yes |
| 16 | GTF2F1 | 1715 | 1.093 | 0.4452 | Yes |
| 17 | NME1 | 1846 | 1.031 | 0.4578 | Yes |
| 18 | POLR1C | 2053 | 0.945 | 0.4645 | Yes |
| 19 | ADA | 2093 | 0.926 | 0.4801 | Yes |
| 20 | MRPL40 | 2171 | 0.892 | 0.4930 | Yes |
| 21 | RAD52 | 2275 | 0.850 | 0.5036 | Yes |
| 22 | GSDME | 2346 | 0.823 | 0.5155 | Yes |
| 23 | DDB2 | 2480 | 0.772 | 0.5229 | Yes |
| 24 | CLP1 | 2497 | 0.766 | 0.5367 | Yes |
| 25 | POLR3GL | 2571 | 0.738 | 0.5469 | Yes |
| 26 | HCLS1 | 2844 | 0.650 | 0.5441 | Yes |
| 27 | SURF1 | 3120 | 0.583 | 0.5400 | Yes |
| 28 | APRT | 3181 | 0.570 | 0.5476 | Yes |
| 29 | NUDT9 | 3231 | 0.556 | 0.5555 | Yes |
| 30 | POLL | 3359 | 0.531 | 0.5587 | Yes |
| 31 | POLR2A | 3369 | 0.529 | 0.5683 | Yes |
| 32 | XPC | 3837 | 0.444 | 0.5507 | Yes |
| 33 | CCNO | 4041 | 0.415 | 0.5474 | Yes |
| 34 | SNAPC4 | 4116 | 0.403 | 0.5510 | Yes |
| 35 | GTF3C5 | 4141 | 0.398 | 0.5573 | Yes |
| 36 | AAAS | 4348 | 0.372 | 0.5529 | Yes |
| 37 | TAF6 | 4357 | 0.372 | 0.5596 | Yes |
| 38 | ELOA | 4369 | 0.370 | 0.5661 | Yes |
| 39 | BRF2 | 4409 | 0.366 | 0.5710 | Yes |
| 40 | GMPR2 | 4492 | 0.356 | 0.5732 | Yes |
| 41 | ADCY6 | 4732 | 0.331 | 0.5662 | No |
| 42 | RFC2 | 4855 | 0.319 | 0.5655 | No |
| 43 | DGUOK | 4862 | 0.319 | 0.5714 | No |
| 44 | POLE4 | 5340 | 0.276 | 0.5500 | No |
| 45 | DUT | 5379 | 0.272 | 0.5531 | No |
| 46 | ERCC3 | 5385 | 0.272 | 0.5580 | No |
| 47 | TAF1C | 5407 | 0.270 | 0.5620 | No |
| 48 | ERCC8 | 5408 | 0.270 | 0.5672 | No |
| 49 | POLR2E | 5757 | 0.245 | 0.5525 | No |
| 50 | ITPA | 5830 | 0.241 | 0.5531 | No |
| 51 | TK2 | 5848 | 0.240 | 0.5567 | No |
| 52 | AK3 | 5966 | 0.233 | 0.5547 | No |
| 53 | RFC4 | 5981 | 0.232 | 0.5584 | No |
| 54 | NT5C3A | 6422 | 0.209 | 0.5377 | No |
| 55 | ARL6IP1 | 6556 | 0.202 | 0.5342 | No |
| 56 | SDCBP | 6653 | 0.198 | 0.5326 | No |
| 57 | STX3 | 6896 | 0.187 | 0.5227 | No |
| 58 | NFX1 | 7107 | 0.178 | 0.5144 | No |
| 59 | POLA2 | 7290 | 0.171 | 0.5075 | No |
| 60 | TAF9 | 7327 | 0.170 | 0.5087 | No |
| 61 | PRIM1 | 7690 | 0.157 | 0.4915 | No |
| 62 | PNP | 7730 | 0.156 | 0.4923 | No |
| 63 | NELFCD | 7767 | 0.155 | 0.4933 | No |
| 64 | POLD1 | 7929 | 0.150 | 0.4871 | No |
| 65 | MPG | 7954 | 0.149 | 0.4887 | No |
| 66 | ERCC1 | 8269 | 0.138 | 0.4737 | No |
| 67 | POLR2C | 8573 | 0.130 | 0.4593 | No |
| 68 | POLR2K | 8926 | 0.121 | 0.4419 | No |
| 69 | GTF2H5 | 8980 | 0.120 | 0.4412 | No |
| 70 | NME4 | 9090 | 0.117 | 0.4374 | No |
| 71 | POLR2J | 9141 | 0.115 | 0.4368 | No |
| 72 | POLR3C | 9161 | 0.115 | 0.4379 | No |
| 73 | GPX4 | 9255 | 0.112 | 0.4349 | No |
| 74 | SUPT4H1 | 9598 | 0.105 | 0.4178 | No |
| 75 | POM121 | 9631 | 0.104 | 0.4180 | No |
| 76 | VPS37B | 9830 | 0.100 | 0.4088 | No |
| 77 | RPA3 | 10018 | 0.097 | 0.4002 | No |
| 78 | POLR2F | 10200 | 0.093 | 0.3919 | No |
| 79 | ZWINT | 10211 | 0.093 | 0.3931 | No |
| 80 | REV3L | 10349 | 0.091 | 0.3872 | No |
| 81 | SAC3D1 | 10359 | 0.091 | 0.3884 | No |
| 82 | NCBP2 | 10413 | 0.090 | 0.3872 | No |
| 83 | POLD3 | 10481 | 0.089 | 0.3851 | No |
| 84 | NME3 | 10848 | 0.082 | 0.3662 | No |
| 85 | CDA | 10857 | 0.082 | 0.3674 | No |
| 86 | RPA2 | 10898 | 0.081 | 0.3667 | No |
| 87 | POLH | 11006 | 0.079 | 0.3622 | No |
| 88 | TAF13 | 11024 | 0.079 | 0.3628 | No |
| 89 | ERCC2 | 11177 | 0.077 | 0.3557 | No |
| 90 | DCTN4 | 11251 | 0.075 | 0.3531 | No |
| 91 | NELFB | 11296 | 0.075 | 0.3521 | No |
| 92 | TYMS | 11417 | 0.073 | 0.3468 | No |
| 93 | SSRP1 | 11447 | 0.072 | 0.3465 | No |
| 94 | TARBP2 | 11464 | 0.072 | 0.3470 | No |
| 95 | UMPS | 11637 | 0.069 | 0.3387 | No |
| 96 | NELFE | 11672 | 0.069 | 0.3381 | No |
| 97 | RNMT | 11755 | 0.068 | 0.3349 | No |
| 98 | MPC2 | 11847 | 0.066 | 0.3310 | No |
| 99 | POLR1D | 11954 | 0.064 | 0.3263 | No |
| 100 | RFC3 | 12285 | 0.060 | 0.3090 | No |
| 101 | AK1 | 12775 | 0.053 | 0.2827 | No |
| 102 | POLR2I | 13013 | 0.050 | 0.2704 | No |
| 103 | RFC5 | 13473 | 0.044 | 0.2455 | No |
| 104 | FEN1 | 13478 | 0.044 | 0.2461 | No |
| 105 | POLR2D | 13699 | 0.041 | 0.2346 | No |
| 106 | GTF2H1 | 13718 | 0.041 | 0.2344 | No |
| 107 | DGCR8 | 13744 | 0.040 | 0.2338 | No |
| 108 | VPS28 | 13885 | 0.038 | 0.2267 | No |
| 109 | SEC61A1 | 13959 | 0.037 | 0.2233 | No |
| 110 | DDB1 | 14048 | 0.035 | 0.2190 | No |
| 111 | CETN2 | 14294 | 0.029 | 0.2059 | No |
| 112 | RALA | 14559 | -0.000 | 0.1911 | No |
| 113 | RAD51 | 14657 | -0.000 | 0.1857 | No |
| 114 | TMED2 | 14815 | -0.000 | 0.1769 | No |
| 115 | RAE1 | 14941 | -0.000 | 0.1699 | No |
| 116 | POLA1 | 14972 | -0.000 | 0.1682 | No |
| 117 | USP11 | 15004 | -0.000 | 0.1665 | No |
| 118 | ELL | 15078 | -0.000 | 0.1624 | No |
| 119 | EDF1 | 15122 | -0.000 | 0.1600 | No |
| 120 | SMAD5 | 15281 | -0.000 | 0.1511 | No |
| 121 | EIF1B | 15299 | -0.000 | 0.1502 | No |
| 122 | TAF12 | 15444 | -0.000 | 0.1421 | No |
| 123 | SRSF6 | 15529 | -0.000 | 0.1374 | No |
| 124 | UPF3B | 15559 | -0.000 | 0.1358 | No |
| 125 | NT5C | 15565 | -0.000 | 0.1355 | No |
| 126 | DAD1 | 15659 | -0.000 | 0.1303 | No |
| 127 | ADRM1 | 15686 | -0.000 | 0.1288 | No |
| 128 | PCNA | 15747 | -0.000 | 0.1255 | No |
| 129 | COX17 | 15912 | -0.000 | 0.1163 | No |
| 130 | GTF2A2 | 15967 | -0.000 | 0.1133 | No |
| 131 | TP53 | 15990 | -0.000 | 0.1120 | No |
| 132 | HPRT1 | 16577 | -0.000 | 0.0792 | No |
| 133 | TAF10 | 16602 | -0.000 | 0.0779 | No |
| 134 | NUDT21 | 16635 | -0.000 | 0.0761 | No |
| 135 | SNAPC5 | 16938 | -0.000 | 0.0592 | No |
| 136 | POLD4 | 16977 | -0.000 | 0.0571 | No |
| 137 | VPS37D | 17002 | -0.000 | 0.0557 | No |
| 138 | SF3A3 | 17190 | -0.000 | 0.0452 | No |
| 139 | ALYREF | 17201 | -0.000 | 0.0447 | No |
| 140 | PDE4B | 17242 | -0.000 | 0.0424 | No |
| 141 | PDE6G | 17272 | -0.000 | 0.0408 | No |
| 142 | BCAP31 | 17292 | -0.000 | 0.0398 | No |
| 143 | SUPT5H | 17431 | -0.000 | 0.0320 | No |