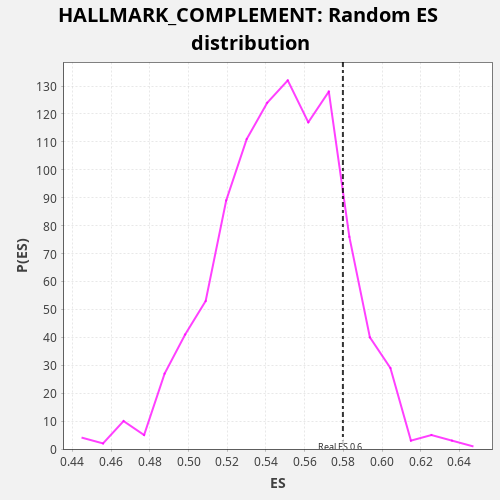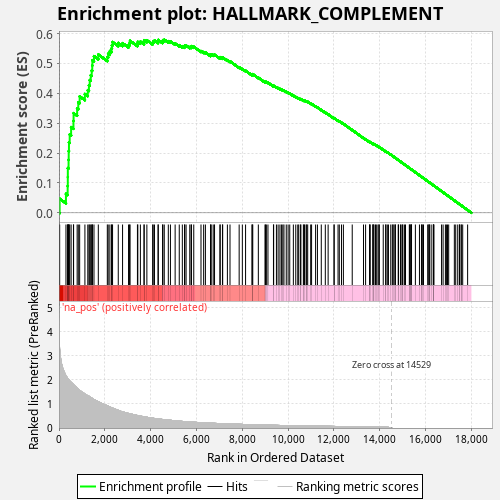
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_COMPLEMENT |
| Enrichment Score (ES) | 0.57979196 |
| Normalized Enrichment Score (NES) | 1.059453 |
| Nominal p-value | 0.141 |
| FDR q-value | 0.6141764 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | JAK2 | 29 | 3.319 | 0.0472 | Yes |
| 2 | LRP1 | 304 | 2.171 | 0.0637 | Yes |
| 3 | MAFF | 378 | 2.060 | 0.0899 | Yes |
| 4 | MMP14 | 384 | 2.049 | 0.1197 | Yes |
| 5 | PSEN1 | 386 | 2.048 | 0.1498 | Yes |
| 6 | S100A12 | 418 | 2.008 | 0.1776 | Yes |
| 7 | RCE1 | 424 | 2.004 | 0.2068 | Yes |
| 8 | C4BPB | 435 | 1.992 | 0.2355 | Yes |
| 9 | F3 | 467 | 1.956 | 0.2625 | Yes |
| 10 | DGKG | 529 | 1.897 | 0.2870 | Yes |
| 11 | MSRB1 | 631 | 1.798 | 0.3077 | Yes |
| 12 | CASP3 | 637 | 1.789 | 0.3337 | Yes |
| 13 | PDP1 | 792 | 1.654 | 0.3494 | Yes |
| 14 | DOCK4 | 842 | 1.610 | 0.3703 | Yes |
| 15 | PLSCR1 | 902 | 1.560 | 0.3900 | Yes |
| 16 | CTSB | 1131 | 1.409 | 0.3979 | Yes |
| 17 | GZMB | 1257 | 1.343 | 0.4106 | Yes |
| 18 | USP14 | 1307 | 1.318 | 0.4272 | Yes |
| 19 | CASP4 | 1341 | 1.295 | 0.4444 | Yes |
| 20 | CPM | 1385 | 1.267 | 0.4606 | Yes |
| 21 | ATOX1 | 1428 | 1.246 | 0.4766 | Yes |
| 22 | PRSS3 | 1452 | 1.226 | 0.4933 | Yes |
| 23 | PLEK | 1458 | 1.222 | 0.5110 | Yes |
| 24 | CTSV | 1527 | 1.183 | 0.5246 | Yes |
| 25 | APOC1 | 1716 | 1.091 | 0.5300 | Yes |
| 26 | DGKH | 2109 | 0.920 | 0.5216 | Yes |
| 27 | GCA | 2147 | 0.906 | 0.5328 | Yes |
| 28 | PIK3R5 | 2220 | 0.873 | 0.5416 | Yes |
| 29 | GNB2 | 2290 | 0.844 | 0.5501 | Yes |
| 30 | IRF2 | 2310 | 0.834 | 0.5613 | Yes |
| 31 | KYNU | 2338 | 0.826 | 0.5719 | Yes |
| 32 | IRF7 | 2587 | 0.732 | 0.5688 | Yes |
| 33 | DOCK10 | 2775 | 0.667 | 0.5681 | Yes |
| 34 | LTF | 3036 | 0.603 | 0.5624 | Yes |
| 35 | SH2B3 | 3082 | 0.592 | 0.5685 | Yes |
| 36 | RABIF | 3101 | 0.586 | 0.5761 | Yes |
| 37 | ADRA2B | 3424 | 0.519 | 0.5657 | Yes |
| 38 | SIRT6 | 3441 | 0.516 | 0.5724 | Yes |
| 39 | CTSC | 3553 | 0.494 | 0.5734 | Yes |
| 40 | KLKB1 | 3714 | 0.463 | 0.5712 | Yes |
| 41 | PLAT | 3716 | 0.462 | 0.5780 | Yes |
| 42 | MMP13 | 3836 | 0.444 | 0.5778 | Yes |
| 43 | F7 | 4088 | 0.407 | 0.5697 | Yes |
| 44 | ERAP2 | 4111 | 0.403 | 0.5744 | Yes |
| 45 | C2 | 4168 | 0.395 | 0.5771 | Yes |
| 46 | TMPRSS6 | 4326 | 0.374 | 0.5737 | Yes |
| 47 | CP | 4333 | 0.373 | 0.5789 | Yes |
| 48 | PCSK9 | 4520 | 0.353 | 0.5736 | Yes |
| 49 | NOTCH4 | 4535 | 0.352 | 0.5780 | Yes |
| 50 | CASP9 | 4595 | 0.345 | 0.5798 | Yes |
| 51 | PSMB9 | 4772 | 0.328 | 0.5747 | No |
| 52 | APOA4 | 4865 | 0.319 | 0.5743 | No |
| 53 | LIPA | 5070 | 0.299 | 0.5672 | No |
| 54 | LAP3 | 5250 | 0.283 | 0.5613 | No |
| 55 | USP16 | 5384 | 0.272 | 0.5578 | No |
| 56 | FN1 | 5479 | 0.265 | 0.5565 | No |
| 57 | ANXA5 | 5491 | 0.264 | 0.5597 | No |
| 58 | CPQ | 5548 | 0.260 | 0.5604 | No |
| 59 | PRCP | 5709 | 0.248 | 0.5551 | No |
| 60 | CASP5 | 5767 | 0.245 | 0.5555 | No |
| 61 | GZMA | 5783 | 0.243 | 0.5582 | No |
| 62 | TFPI2 | 5883 | 0.237 | 0.5561 | No |
| 63 | CCL5 | 6199 | 0.221 | 0.5417 | No |
| 64 | MMP12 | 6315 | 0.215 | 0.5384 | No |
| 65 | EHD1 | 6391 | 0.211 | 0.5373 | No |
| 66 | CTSO | 6614 | 0.199 | 0.5278 | No |
| 67 | CD36 | 6622 | 0.199 | 0.5303 | No |
| 68 | CD55 | 6672 | 0.197 | 0.5304 | No |
| 69 | PRDM4 | 6758 | 0.193 | 0.5285 | No |
| 70 | LGALS3 | 6790 | 0.191 | 0.5296 | No |
| 71 | KLK1 | 7027 | 0.181 | 0.5190 | No |
| 72 | SCG3 | 7036 | 0.181 | 0.5212 | No |
| 73 | CFB | 7136 | 0.177 | 0.5182 | No |
| 74 | LCK | 7146 | 0.176 | 0.5203 | No |
| 75 | C9 | 7351 | 0.169 | 0.5113 | No |
| 76 | SERPINB2 | 7465 | 0.165 | 0.5074 | No |
| 77 | LCP2 | 7865 | 0.152 | 0.4873 | No |
| 78 | COL4A2 | 8004 | 0.147 | 0.4817 | No |
| 79 | FCN1 | 8145 | 0.142 | 0.4759 | No |
| 80 | DOCK9 | 8429 | 0.134 | 0.4620 | No |
| 81 | PRSS36 | 8433 | 0.134 | 0.4638 | No |
| 82 | PLG | 8459 | 0.133 | 0.4643 | No |
| 83 | RBSN | 8702 | 0.127 | 0.4526 | No |
| 84 | GP1BA | 8993 | 0.119 | 0.4381 | No |
| 85 | CASP10 | 9015 | 0.119 | 0.4387 | No |
| 86 | PREP | 9053 | 0.118 | 0.4383 | No |
| 87 | GZMK | 9122 | 0.116 | 0.4362 | No |
| 88 | OLR1 | 9363 | 0.110 | 0.4243 | No |
| 89 | MMP8 | 9371 | 0.110 | 0.4256 | No |
| 90 | CFH | 9492 | 0.107 | 0.4204 | No |
| 91 | CR1 | 9565 | 0.106 | 0.4179 | No |
| 92 | LGMN | 9645 | 0.104 | 0.4150 | No |
| 93 | APOBEC3F | 9710 | 0.103 | 0.4129 | No |
| 94 | ITGAM | 9770 | 0.101 | 0.4111 | No |
| 95 | ITIH1 | 9820 | 0.100 | 0.4098 | No |
| 96 | STX4 | 9921 | 0.098 | 0.4057 | No |
| 97 | CBLB | 10002 | 0.097 | 0.4026 | No |
| 98 | GNAI3 | 10073 | 0.096 | 0.4001 | No |
| 99 | FDX1 | 10231 | 0.093 | 0.3926 | No |
| 100 | F2 | 10336 | 0.091 | 0.3881 | No |
| 101 | DUSP5 | 10424 | 0.090 | 0.3845 | No |
| 102 | CTSL | 10464 | 0.089 | 0.3837 | No |
| 103 | DPP4 | 10539 | 0.088 | 0.3808 | No |
| 104 | RASGRP1 | 10560 | 0.087 | 0.3810 | No |
| 105 | GNB4 | 10668 | 0.085 | 0.3762 | No |
| 106 | F10 | 10693 | 0.085 | 0.3761 | No |
| 107 | KCNIP3 | 10719 | 0.084 | 0.3759 | No |
| 108 | SERPINA1 | 10787 | 0.083 | 0.3734 | No |
| 109 | PCLO | 10813 | 0.082 | 0.3732 | No |
| 110 | CDA | 10857 | 0.082 | 0.3720 | No |
| 111 | ACTN2 | 10988 | 0.079 | 0.3659 | No |
| 112 | MMP15 | 11027 | 0.079 | 0.3649 | No |
| 113 | CASP1 | 11202 | 0.076 | 0.3562 | No |
| 114 | APOBEC3G | 11282 | 0.075 | 0.3529 | No |
| 115 | PRKCD | 11450 | 0.072 | 0.3446 | No |
| 116 | CDH13 | 11626 | 0.070 | 0.3358 | No |
| 117 | XPNPEP1 | 11749 | 0.068 | 0.3299 | No |
| 118 | PLAUR | 12009 | 0.064 | 0.3163 | No |
| 119 | CLU | 12010 | 0.064 | 0.3173 | No |
| 120 | CTSS | 12182 | 0.061 | 0.3086 | No |
| 121 | PLA2G4A | 12237 | 0.061 | 0.3064 | No |
| 122 | CA2 | 12332 | 0.060 | 0.3020 | No |
| 123 | SPOCK2 | 12415 | 0.058 | 0.2983 | No |
| 124 | CTSH | 12801 | 0.053 | 0.2774 | No |
| 125 | CXCL1 | 13293 | 0.047 | 0.2506 | No |
| 126 | BRPF3 | 13385 | 0.045 | 0.2461 | No |
| 127 | HPCAL4 | 13562 | 0.042 | 0.2369 | No |
| 128 | S100A9 | 13568 | 0.042 | 0.2372 | No |
| 129 | AKAP10 | 13581 | 0.042 | 0.2371 | No |
| 130 | CD40LG | 13704 | 0.041 | 0.2309 | No |
| 131 | PIK3CA | 13712 | 0.041 | 0.2311 | No |
| 132 | HSPA5 | 13713 | 0.041 | 0.2317 | No |
| 133 | CASP7 | 13751 | 0.040 | 0.2302 | No |
| 134 | KCNIP2 | 13817 | 0.039 | 0.2271 | No |
| 135 | PPP2CB | 13852 | 0.039 | 0.2258 | No |
| 136 | C3 | 13925 | 0.037 | 0.2223 | No |
| 137 | DYRK2 | 13965 | 0.037 | 0.2207 | No |
| 138 | DUSP6 | 13985 | 0.036 | 0.2201 | No |
| 139 | KIF2A | 14155 | 0.033 | 0.2111 | No |
| 140 | PFN1 | 14264 | 0.030 | 0.2055 | No |
| 141 | GRB2 | 14267 | 0.030 | 0.2058 | No |
| 142 | WAS | 14332 | 0.027 | 0.2026 | No |
| 143 | GNGT2 | 14383 | 0.023 | 0.2002 | No |
| 144 | RHOG | 14483 | 0.017 | 0.1949 | No |
| 145 | LAMP2 | 14552 | -0.000 | 0.1911 | No |
| 146 | FYN | 14583 | -0.000 | 0.1894 | No |
| 147 | TIMP2 | 14627 | -0.000 | 0.1870 | No |
| 148 | RNF4 | 14678 | -0.000 | 0.1842 | No |
| 149 | ME1 | 14685 | -0.000 | 0.1838 | No |
| 150 | CD59 | 14809 | -0.000 | 0.1769 | No |
| 151 | MT3 | 14822 | -0.000 | 0.1762 | No |
| 152 | PDGFB | 14907 | -0.000 | 0.1715 | No |
| 153 | HNF4A | 14935 | -0.000 | 0.1700 | No |
| 154 | PHEX | 15001 | -0.000 | 0.1664 | No |
| 155 | TIMP1 | 15009 | -0.000 | 0.1660 | No |
| 156 | PIK3CG | 15087 | -0.000 | 0.1616 | No |
| 157 | GATA3 | 15127 | -0.000 | 0.1595 | No |
| 158 | GNAI2 | 15294 | -0.000 | 0.1501 | No |
| 159 | GPD2 | 15307 | -0.000 | 0.1495 | No |
| 160 | CR2 | 15362 | -0.000 | 0.1464 | No |
| 161 | CD46 | 15363 | -0.000 | 0.1464 | No |
| 162 | SERPINC1 | 15374 | -0.000 | 0.1459 | No |
| 163 | TNFAIP3 | 15390 | -0.000 | 0.1450 | No |
| 164 | IRF1 | 15558 | -0.000 | 0.1356 | No |
| 165 | RAF1 | 15736 | -0.000 | 0.1257 | No |
| 166 | USP15 | 15824 | -0.000 | 0.1208 | No |
| 167 | IL6 | 15843 | -0.000 | 0.1198 | No |
| 168 | PIM1 | 15881 | -0.000 | 0.1177 | No |
| 169 | USP8 | 15913 | -0.000 | 0.1160 | No |
| 170 | PLA2G7 | 16096 | -0.000 | 0.1058 | No |
| 171 | ZEB1 | 16148 | -0.000 | 0.1029 | No |
| 172 | SERPING1 | 16156 | -0.000 | 0.1025 | No |
| 173 | PPP4C | 16167 | -0.000 | 0.1020 | No |
| 174 | L3MBTL4 | 16247 | -0.000 | 0.0975 | No |
| 175 | FCER1G | 16339 | -0.000 | 0.0924 | No |
| 176 | C1QC | 16346 | -0.000 | 0.0921 | No |
| 177 | C1R | 16355 | -0.000 | 0.0916 | No |
| 178 | CALM3 | 16366 | -0.000 | 0.0911 | No |
| 179 | ADAM9 | 16701 | -0.000 | 0.0723 | No |
| 180 | ZFPM2 | 16768 | -0.000 | 0.0686 | No |
| 181 | CEBPB | 16869 | -0.000 | 0.0630 | No |
| 182 | C1QA | 16913 | -0.000 | 0.0606 | No |
| 183 | VCPIP1 | 16961 | -0.000 | 0.0579 | No |
| 184 | CDK5R1 | 17012 | -0.000 | 0.0551 | No |
| 185 | F8 | 17259 | -0.000 | 0.0413 | No |
| 186 | GNG2 | 17319 | -0.000 | 0.0380 | No |
| 187 | S100A13 | 17407 | -0.000 | 0.0331 | No |
| 188 | GMFB | 17480 | -0.000 | 0.0291 | No |
| 189 | SRC | 17485 | -0.000 | 0.0289 | No |
| 190 | CALM1 | 17557 | -0.000 | 0.0249 | No |
| 191 | F5 | 17561 | -0.000 | 0.0247 | No |
| 192 | HSPA1A | 17624 | -0.000 | 0.0212 | No |
| 193 | LYN | 17839 | -0.000 | 0.0092 | No |