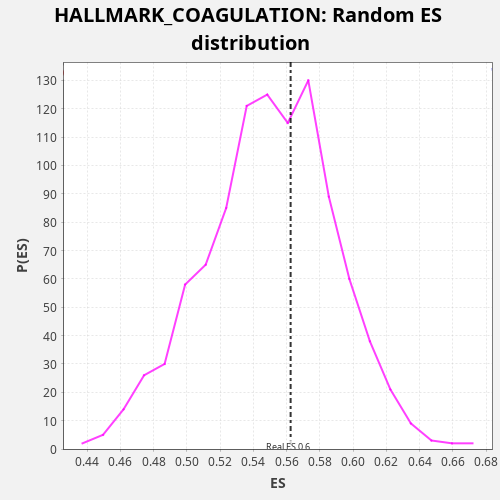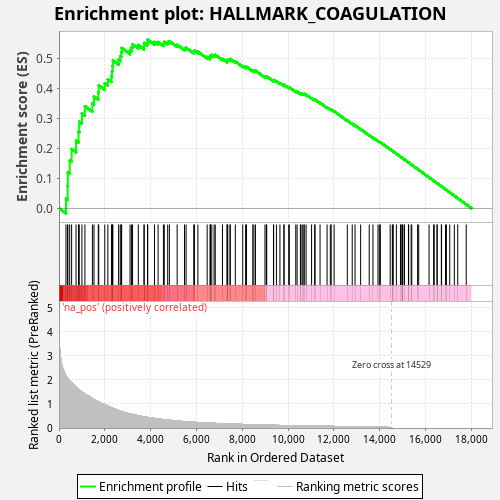
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | 0.56238997 |
| Normalized Enrichment Score (NES) | 1.021882 |
| Nominal p-value | 0.401 |
| FDR q-value | 0.7814801 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | LRP1 | 304 | 2.171 | 0.0319 | Yes |
| 2 | MAFF | 378 | 2.060 | 0.0743 | Yes |
| 3 | MMP14 | 384 | 2.049 | 0.1202 | Yes |
| 4 | F3 | 467 | 1.956 | 0.1597 | Yes |
| 5 | FURIN | 551 | 1.878 | 0.1974 | Yes |
| 6 | PEF1 | 744 | 1.701 | 0.2249 | Yes |
| 7 | MSRB2 | 851 | 1.600 | 0.2551 | Yes |
| 8 | TIMP3 | 884 | 1.573 | 0.2887 | Yes |
| 9 | APOA1 | 1001 | 1.494 | 0.3159 | Yes |
| 10 | CTSB | 1131 | 1.409 | 0.3404 | Yes |
| 11 | PLEK | 1458 | 1.222 | 0.3498 | Yes |
| 12 | CTSV | 1527 | 1.183 | 0.3726 | Yes |
| 13 | APOC1 | 1716 | 1.091 | 0.3867 | Yes |
| 14 | GSN | 1735 | 1.080 | 0.4100 | Yes |
| 15 | ACOX2 | 1998 | 0.964 | 0.4171 | Yes |
| 16 | A2M | 2133 | 0.911 | 0.4301 | Yes |
| 17 | GNB2 | 2290 | 0.844 | 0.4404 | Yes |
| 18 | CRIP2 | 2314 | 0.833 | 0.4579 | Yes |
| 19 | CPB2 | 2336 | 0.827 | 0.4754 | Yes |
| 20 | ANXA1 | 2351 | 0.821 | 0.4931 | Yes |
| 21 | F13B | 2603 | 0.727 | 0.4954 | Yes |
| 22 | C8G | 2674 | 0.700 | 0.5073 | Yes |
| 23 | CAPN5 | 2718 | 0.685 | 0.5203 | Yes |
| 24 | CAPN2 | 2735 | 0.680 | 0.5348 | Yes |
| 25 | RABIF | 3101 | 0.586 | 0.5275 | Yes |
| 26 | ITGB3 | 3163 | 0.573 | 0.5370 | Yes |
| 27 | BMP1 | 3211 | 0.560 | 0.5470 | Yes |
| 28 | MMP10 | 3462 | 0.512 | 0.5446 | Yes |
| 29 | KLKB1 | 3714 | 0.463 | 0.5410 | Yes |
| 30 | PLAT | 3716 | 0.462 | 0.5514 | Yes |
| 31 | F11 | 3864 | 0.440 | 0.5530 | Yes |
| 32 | CPN1 | 3874 | 0.437 | 0.5624 | Yes |
| 33 | C2 | 4168 | 0.395 | 0.5549 | No |
| 34 | TMPRSS6 | 4326 | 0.374 | 0.5545 | No |
| 35 | CTSE | 4559 | 0.349 | 0.5494 | No |
| 36 | CASP9 | 4595 | 0.345 | 0.5552 | No |
| 37 | HTRA1 | 4745 | 0.330 | 0.5543 | No |
| 38 | PROZ | 4818 | 0.323 | 0.5576 | No |
| 39 | CFD | 5158 | 0.292 | 0.5452 | No |
| 40 | FN1 | 5479 | 0.265 | 0.5333 | No |
| 41 | CPQ | 5548 | 0.260 | 0.5353 | No |
| 42 | TFPI2 | 5883 | 0.237 | 0.5220 | No |
| 43 | MMP11 | 5914 | 0.236 | 0.5256 | No |
| 44 | CTSK | 6063 | 0.228 | 0.5225 | No |
| 45 | MMP3 | 6468 | 0.207 | 0.5045 | No |
| 46 | RAPGEF3 | 6593 | 0.201 | 0.5021 | No |
| 47 | PF4 | 6613 | 0.200 | 0.5056 | No |
| 48 | CTSO | 6614 | 0.199 | 0.5101 | No |
| 49 | HRG | 6671 | 0.197 | 0.5114 | No |
| 50 | COMP | 6771 | 0.192 | 0.5102 | No |
| 51 | RGN | 6821 | 0.190 | 0.5117 | No |
| 52 | CFB | 7136 | 0.177 | 0.4981 | No |
| 53 | FGA | 7334 | 0.170 | 0.4909 | No |
| 54 | C9 | 7351 | 0.169 | 0.4938 | No |
| 55 | CFI | 7367 | 0.169 | 0.4968 | No |
| 56 | SERPINB2 | 7465 | 0.165 | 0.4951 | No |
| 57 | TF | 7479 | 0.165 | 0.4981 | No |
| 58 | LEFTY2 | 7697 | 0.157 | 0.4895 | No |
| 59 | PLAU | 8021 | 0.147 | 0.4747 | No |
| 60 | VWF | 8147 | 0.142 | 0.4709 | No |
| 61 | SH2B2 | 8186 | 0.141 | 0.4720 | No |
| 62 | PLG | 8459 | 0.133 | 0.4598 | No |
| 63 | MMP7 | 8509 | 0.132 | 0.4600 | No |
| 64 | C8A | 8576 | 0.130 | 0.4592 | No |
| 65 | GP1BA | 8993 | 0.119 | 0.4386 | No |
| 66 | PREP | 9053 | 0.118 | 0.4380 | No |
| 67 | RAC1 | 9068 | 0.117 | 0.4399 | No |
| 68 | OLR1 | 9363 | 0.110 | 0.4259 | No |
| 69 | MMP8 | 9371 | 0.110 | 0.4280 | No |
| 70 | CFH | 9492 | 0.107 | 0.4237 | No |
| 71 | LGMN | 9645 | 0.104 | 0.4175 | No |
| 72 | ITIH1 | 9820 | 0.100 | 0.4100 | No |
| 73 | DCT | 9826 | 0.100 | 0.4120 | No |
| 74 | MMP1 | 10029 | 0.096 | 0.4029 | No |
| 75 | MST1 | 10053 | 0.096 | 0.4038 | No |
| 76 | F2 | 10336 | 0.091 | 0.3900 | No |
| 77 | APOC3 | 10394 | 0.090 | 0.3889 | No |
| 78 | DPP4 | 10539 | 0.088 | 0.3828 | No |
| 79 | PRSS23 | 10561 | 0.087 | 0.3836 | No |
| 80 | MMP2 | 10620 | 0.086 | 0.3823 | No |
| 81 | F10 | 10693 | 0.085 | 0.3802 | No |
| 82 | HMGCS2 | 10704 | 0.085 | 0.3815 | No |
| 83 | SERPINA1 | 10787 | 0.083 | 0.3788 | No |
| 84 | MMP15 | 11027 | 0.079 | 0.3672 | No |
| 85 | MMP9 | 11161 | 0.077 | 0.3615 | No |
| 86 | THBS1 | 11178 | 0.077 | 0.3623 | No |
| 87 | MASP2 | 11396 | 0.073 | 0.3518 | No |
| 88 | C8B | 11703 | 0.068 | 0.3362 | No |
| 89 | PROS1 | 11844 | 0.066 | 0.3299 | No |
| 90 | ISCU | 11883 | 0.066 | 0.3292 | No |
| 91 | CLU | 12010 | 0.064 | 0.3236 | No |
| 92 | P2RY1 | 12587 | 0.056 | 0.2927 | No |
| 93 | CTSH | 12801 | 0.053 | 0.2819 | No |
| 94 | PROC | 12924 | 0.051 | 0.2763 | No |
| 95 | MBL2 | 13168 | 0.048 | 0.2638 | No |
| 96 | FGG | 13543 | 0.043 | 0.2438 | No |
| 97 | FBN1 | 13706 | 0.041 | 0.2357 | No |
| 98 | C3 | 13925 | 0.037 | 0.2243 | No |
| 99 | DUSP6 | 13985 | 0.036 | 0.2218 | No |
| 100 | DUSP14 | 14028 | 0.036 | 0.2203 | No |
| 101 | KLF7 | 14458 | 0.019 | 0.1967 | No |
| 102 | LAMP2 | 14552 | -0.000 | 0.1915 | No |
| 103 | CD9 | 14579 | -0.000 | 0.1901 | No |
| 104 | FYN | 14583 | -0.000 | 0.1899 | No |
| 105 | WDR1 | 14729 | -0.000 | 0.1818 | No |
| 106 | PDGFB | 14907 | -0.000 | 0.1719 | No |
| 107 | HNF4A | 14935 | -0.000 | 0.1704 | No |
| 108 | F9 | 14983 | -0.000 | 0.1677 | No |
| 109 | USP11 | 15004 | -0.000 | 0.1666 | No |
| 110 | TIMP1 | 15009 | -0.000 | 0.1664 | No |
| 111 | HPN | 15082 | -0.000 | 0.1624 | No |
| 112 | MEP1A | 15256 | -0.000 | 0.1527 | No |
| 113 | SPARC | 15265 | -0.000 | 0.1522 | No |
| 114 | SERPINC1 | 15374 | -0.000 | 0.1462 | No |
| 115 | GDA | 15400 | -0.000 | 0.1448 | No |
| 116 | KLK8 | 15655 | -0.000 | 0.1306 | No |
| 117 | F12 | 15703 | -0.000 | 0.1280 | No |
| 118 | SERPING1 | 16156 | -0.000 | 0.1027 | No |
| 119 | C1R | 16355 | -0.000 | 0.0916 | No |
| 120 | S100A1 | 16380 | -0.000 | 0.0902 | No |
| 121 | ITGA2 | 16506 | -0.000 | 0.0832 | No |
| 122 | F2RL2 | 16508 | -0.000 | 0.0832 | No |
| 123 | ARF4 | 16692 | -0.000 | 0.0730 | No |
| 124 | ADAM9 | 16701 | -0.000 | 0.0725 | No |
| 125 | GNG12 | 16874 | -0.000 | 0.0629 | No |
| 126 | C1QA | 16913 | -0.000 | 0.0608 | No |
| 127 | THBD | 17058 | -0.000 | 0.0527 | No |
| 128 | F8 | 17259 | -0.000 | 0.0415 | No |
| 129 | S100A13 | 17407 | -0.000 | 0.0333 | No |
| 130 | APOC2 | 17778 | -0.000 | 0.0126 | No |