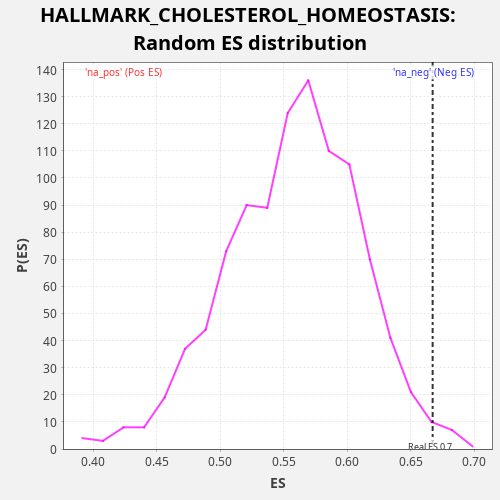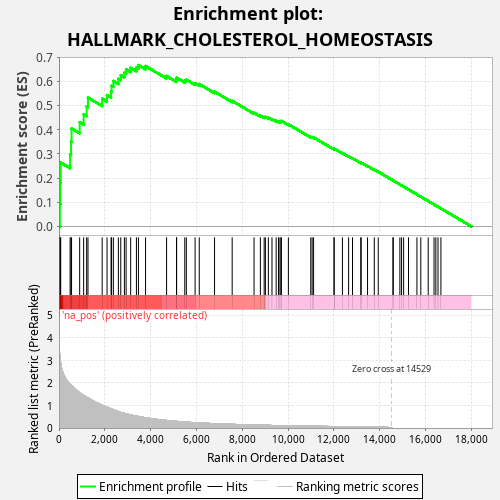
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | 0.66715926 |
| Normalized Enrichment Score (NES) | 1.196564 |
| Nominal p-value | 0.014 |
| FDR q-value | 0.11025004 |
| FWER p-Value | 0.36 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | ABCA2 | 37 | 3.230 | 0.0926 | Yes |
| 2 | ATF5 | 58 | 3.017 | 0.1799 | Yes |
| 3 | FDFT1 | 75 | 2.893 | 0.2638 | Yes |
| 4 | IDI1 | 485 | 1.934 | 0.2977 | Yes |
| 5 | GLDC | 525 | 1.899 | 0.3511 | Yes |
| 6 | PDK3 | 548 | 1.880 | 0.4050 | Yes |
| 7 | PLSCR1 | 902 | 1.560 | 0.4310 | Yes |
| 8 | CXCL16 | 1076 | 1.448 | 0.4638 | Yes |
| 9 | GUSB | 1208 | 1.363 | 0.4965 | Yes |
| 10 | CHKA | 1265 | 1.338 | 0.5325 | Yes |
| 11 | ADH4 | 1885 | 1.013 | 0.5277 | Yes |
| 12 | LDLR | 2097 | 0.924 | 0.5430 | Yes |
| 13 | SREBF2 | 2273 | 0.851 | 0.5582 | Yes |
| 14 | AVPR1A | 2295 | 0.841 | 0.5817 | Yes |
| 15 | CYP51A1 | 2373 | 0.814 | 0.6012 | Yes |
| 16 | MVK | 2592 | 0.731 | 0.6105 | Yes |
| 17 | LSS | 2696 | 0.693 | 0.6251 | Yes |
| 18 | ACSS2 | 2855 | 0.648 | 0.6353 | Yes |
| 19 | CPEB2 | 2930 | 0.629 | 0.6496 | Yes |
| 20 | SC5D | 3129 | 0.580 | 0.6555 | Yes |
| 21 | STARD4 | 3380 | 0.528 | 0.6571 | Yes |
| 22 | DHCR7 | 3468 | 0.510 | 0.6672 | Yes |
| 23 | TRIB3 | 3779 | 0.453 | 0.6631 | No |
| 24 | MVD | 4697 | 0.334 | 0.6218 | No |
| 25 | ERRFI1 | 5130 | 0.294 | 0.6063 | No |
| 26 | ANXA13 | 5135 | 0.294 | 0.6147 | No |
| 27 | ANXA5 | 5491 | 0.264 | 0.6027 | No |
| 28 | ACAT2 | 5559 | 0.260 | 0.6065 | No |
| 29 | TM7SF2 | 5941 | 0.234 | 0.5922 | No |
| 30 | GSTM2 | 6123 | 0.225 | 0.5887 | No |
| 31 | LGALS3 | 6790 | 0.191 | 0.5571 | No |
| 32 | LPL | 7560 | 0.162 | 0.5190 | No |
| 33 | TMEM97 | 8515 | 0.132 | 0.4696 | No |
| 34 | PCYT2 | 8791 | 0.125 | 0.4580 | No |
| 35 | GPX8 | 8957 | 0.120 | 0.4523 | No |
| 36 | HSD17B7 | 9011 | 0.119 | 0.4528 | No |
| 37 | TNFRSF12A | 9140 | 0.115 | 0.4490 | No |
| 38 | ALDOC | 9298 | 0.112 | 0.4436 | No |
| 39 | MAL2 | 9480 | 0.108 | 0.4366 | No |
| 40 | ETHE1 | 9582 | 0.106 | 0.4341 | No |
| 41 | LGMN | 9645 | 0.104 | 0.4337 | No |
| 42 | ATXN2 | 9691 | 0.103 | 0.4342 | No |
| 43 | ANTXR2 | 9707 | 0.103 | 0.4364 | No |
| 44 | NIBAN1 | 10014 | 0.097 | 0.4221 | No |
| 45 | HMGCR | 10991 | 0.079 | 0.3700 | No |
| 46 | FDPS | 11051 | 0.078 | 0.3690 | No |
| 47 | TP53INP1 | 11114 | 0.077 | 0.3679 | No |
| 48 | PLAUR | 12009 | 0.064 | 0.3199 | No |
| 49 | CLU | 12010 | 0.064 | 0.3217 | No |
| 50 | ALCAM | 12373 | 0.059 | 0.3033 | No |
| 51 | PNRC1 | 12643 | 0.055 | 0.2899 | No |
| 52 | ATF3 | 12813 | 0.053 | 0.2820 | No |
| 53 | FABP5 | 13175 | 0.048 | 0.2633 | No |
| 54 | FBXO6 | 13190 | 0.048 | 0.2639 | No |
| 55 | FASN | 13470 | 0.044 | 0.2497 | No |
| 56 | ACTG1 | 13764 | 0.040 | 0.2345 | No |
| 57 | PPARG | 13937 | 0.037 | 0.2260 | No |
| 58 | CD9 | 14579 | -0.000 | 0.1903 | No |
| 59 | SEMA3B | 14591 | -0.000 | 0.1896 | No |
| 60 | SCD | 14881 | -0.000 | 0.1735 | No |
| 61 | JAG1 | 14955 | -0.000 | 0.1695 | No |
| 62 | SQLE | 15045 | -0.000 | 0.1645 | No |
| 63 | HMGCS1 | 15258 | -0.000 | 0.1527 | No |
| 64 | GNAI1 | 15626 | -0.000 | 0.1322 | No |
| 65 | FADS2 | 15795 | -0.000 | 0.1228 | No |
| 66 | EBP | 16118 | -0.000 | 0.1049 | No |
| 67 | CBS | 16372 | -0.000 | 0.0908 | No |
| 68 | PMVK | 16451 | -0.000 | 0.0864 | No |
| 69 | NFIL3 | 16546 | -0.000 | 0.0812 | No |
| 70 | CTNNB1 | 16673 | -0.000 | 0.0742 | No |