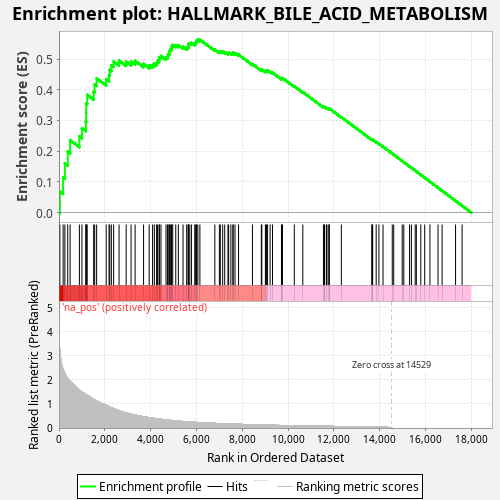
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
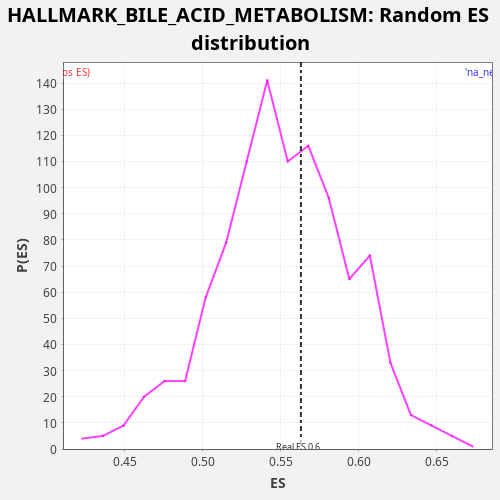
| Enrichment Score (ES) | 0.5629724 |
| Normalized Enrichment Score (NES) | 1.0214912 |
| Nominal p-value | 0.4 |
| FDR q-value | 0.7554616 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | ABCA2 | 37 | 3.230 | 0.0692 | Yes |
| 2 | PAOX | 180 | 2.440 | 0.1151 | Yes |
| 3 | NR1H4 | 256 | 2.253 | 0.1606 | Yes |
| 4 | BMP6 | 381 | 2.058 | 0.1990 | Yes |
| 5 | IDI1 | 485 | 1.934 | 0.2359 | Yes |
| 6 | FADS1 | 890 | 1.571 | 0.2480 | Yes |
| 7 | APOA1 | 1001 | 1.494 | 0.2748 | Yes |
| 8 | DIO1 | 1172 | 1.389 | 0.2960 | Yes |
| 9 | OPTN | 1187 | 1.377 | 0.3255 | Yes |
| 10 | MLYCD | 1189 | 1.376 | 0.3558 | Yes |
| 11 | IDH2 | 1231 | 1.354 | 0.3834 | Yes |
| 12 | PXMP2 | 1513 | 1.187 | 0.3939 | Yes |
| 13 | SLC35B2 | 1558 | 1.166 | 0.4172 | Yes |
| 14 | GNMT | 1643 | 1.120 | 0.4372 | Yes |
| 15 | CYP7A1 | 2060 | 0.941 | 0.4347 | Yes |
| 16 | ABCD3 | 2185 | 0.885 | 0.4473 | Yes |
| 17 | CYP39A1 | 2215 | 0.874 | 0.4649 | Yes |
| 18 | ABCA6 | 2285 | 0.845 | 0.4797 | Yes |
| 19 | SLC27A5 | 2385 | 0.810 | 0.4920 | Yes |
| 20 | NEDD4 | 2622 | 0.720 | 0.4947 | Yes |
| 21 | ABCA8 | 2932 | 0.629 | 0.4913 | Yes |
| 22 | PEX7 | 3146 | 0.576 | 0.4921 | Yes |
| 23 | CAT | 3323 | 0.539 | 0.4942 | Yes |
| 24 | EFHC1 | 3692 | 0.467 | 0.4839 | Yes |
| 25 | CYP27A1 | 3936 | 0.428 | 0.4798 | Yes |
| 26 | AKR1D1 | 4085 | 0.407 | 0.4805 | Yes |
| 27 | ABCG8 | 4162 | 0.395 | 0.4850 | Yes |
| 28 | HSD17B4 | 4261 | 0.382 | 0.4879 | Yes |
| 29 | PHYH | 4303 | 0.377 | 0.4940 | Yes |
| 30 | ABCA9 | 4371 | 0.370 | 0.4984 | Yes |
| 31 | ACSL5 | 4381 | 0.369 | 0.5060 | Yes |
| 32 | PEX12 | 4451 | 0.361 | 0.5101 | Yes |
| 33 | GSTK1 | 4672 | 0.338 | 0.5053 | Yes |
| 34 | HSD3B7 | 4742 | 0.330 | 0.5087 | Yes |
| 35 | SOAT2 | 4756 | 0.329 | 0.5152 | Yes |
| 36 | SLCO1A2 | 4810 | 0.324 | 0.5194 | Yes |
| 37 | CH25H | 4815 | 0.323 | 0.5263 | Yes |
| 38 | KLF1 | 4860 | 0.319 | 0.5309 | Yes |
| 39 | AGXT | 4910 | 0.314 | 0.5351 | Yes |
| 40 | ABCA1 | 4919 | 0.313 | 0.5415 | Yes |
| 41 | BCAR3 | 4965 | 0.309 | 0.5458 | Yes |
| 42 | PEX6 | 5097 | 0.298 | 0.5451 | Yes |
| 43 | SULT1B1 | 5211 | 0.287 | 0.5451 | Yes |
| 44 | HAO1 | 5415 | 0.270 | 0.5397 | Yes |
| 45 | LONP2 | 5574 | 0.258 | 0.5366 | Yes |
| 46 | FDXR | 5638 | 0.253 | 0.5386 | Yes |
| 47 | NUDT12 | 5650 | 0.252 | 0.5436 | Yes |
| 48 | PFKM | 5652 | 0.252 | 0.5491 | Yes |
| 49 | LIPE | 5708 | 0.248 | 0.5515 | Yes |
| 50 | EPHX2 | 5781 | 0.244 | 0.5528 | Yes |
| 51 | NR1I2 | 5927 | 0.235 | 0.5499 | Yes |
| 52 | SLC22A18 | 5968 | 0.233 | 0.5528 | Yes |
| 53 | BBOX1 | 5995 | 0.232 | 0.5565 | Yes |
| 54 | PNPLA8 | 6009 | 0.231 | 0.5608 | Yes |
| 55 | PEX11A | 6062 | 0.228 | 0.5630 | Yes |
| 56 | CYP7B1 | 6151 | 0.224 | 0.5630 | Yes |
| 57 | IDH1 | 6803 | 0.191 | 0.5308 | No |
| 58 | ABCA4 | 7001 | 0.182 | 0.5238 | No |
| 59 | RETSAT | 7043 | 0.181 | 0.5255 | No |
| 60 | LCK | 7146 | 0.176 | 0.5237 | No |
| 61 | GC | 7234 | 0.173 | 0.5227 | No |
| 62 | PEX26 | 7376 | 0.168 | 0.5185 | No |
| 63 | NR0B2 | 7406 | 0.167 | 0.5205 | No |
| 64 | ABCD2 | 7503 | 0.164 | 0.5188 | No |
| 65 | PECR | 7594 | 0.161 | 0.5173 | No |
| 66 | ALDH8A1 | 7600 | 0.160 | 0.5206 | No |
| 67 | PEX19 | 7684 | 0.157 | 0.5194 | No |
| 68 | ABCG4 | 7838 | 0.153 | 0.5142 | No |
| 69 | ISOC1 | 8445 | 0.133 | 0.4833 | No |
| 70 | PEX1 | 8832 | 0.124 | 0.4644 | No |
| 71 | SULT2B1 | 8855 | 0.123 | 0.4659 | No |
| 72 | PEX13 | 9010 | 0.119 | 0.4599 | No |
| 73 | PIPOX | 9037 | 0.118 | 0.4611 | No |
| 74 | HACL1 | 9054 | 0.118 | 0.4628 | No |
| 75 | SCP2 | 9105 | 0.117 | 0.4626 | No |
| 76 | ABCA5 | 9217 | 0.113 | 0.4589 | No |
| 77 | GNPAT | 9321 | 0.111 | 0.4556 | No |
| 78 | AQP9 | 9712 | 0.103 | 0.4360 | No |
| 79 | PEX16 | 9733 | 0.102 | 0.4372 | No |
| 80 | AMACR | 9755 | 0.102 | 0.4382 | No |
| 81 | ALDH9A1 | 10273 | 0.092 | 0.4114 | No |
| 82 | SLC29A1 | 10644 | 0.086 | 0.3926 | No |
| 83 | DHCR24 | 11545 | 0.071 | 0.3439 | No |
| 84 | SLC23A1 | 11592 | 0.070 | 0.3428 | No |
| 85 | SLC23A2 | 11597 | 0.070 | 0.3442 | No |
| 86 | ACSL1 | 11679 | 0.069 | 0.3411 | No |
| 87 | SOD1 | 11745 | 0.068 | 0.3390 | No |
| 88 | RBP1 | 11793 | 0.067 | 0.3379 | No |
| 89 | SERPINA6 | 11797 | 0.067 | 0.3392 | No |
| 90 | HSD17B11 | 12326 | 0.060 | 0.3110 | No |
| 91 | DIO2 | 13653 | 0.042 | 0.2378 | No |
| 92 | GCLM | 13683 | 0.041 | 0.2371 | No |
| 93 | RXRG | 13693 | 0.041 | 0.2375 | No |
| 94 | CYP46A1 | 13847 | 0.039 | 0.2298 | No |
| 95 | NPC1 | 13967 | 0.037 | 0.2239 | No |
| 96 | CYP8B1 | 14146 | 0.033 | 0.2147 | No |
| 97 | CROT | 14549 | -0.000 | 0.1923 | No |
| 98 | HSD17B6 | 14612 | -0.000 | 0.1888 | No |
| 99 | ABCD1 | 14984 | -0.000 | 0.1681 | No |
| 100 | PEX11G | 15053 | -0.000 | 0.1643 | No |
| 101 | TFCP2L1 | 15304 | -0.000 | 0.1503 | No |
| 102 | TTR | 15385 | -0.000 | 0.1458 | No |
| 103 | ATXN1 | 15550 | -0.000 | 0.1366 | No |
| 104 | PRDX5 | 15600 | -0.000 | 0.1339 | No |
| 105 | FADS2 | 15795 | -0.000 | 0.1231 | No |
| 106 | SLC27A2 | 15965 | -0.000 | 0.1136 | No |
| 107 | NR3C2 | 16194 | -0.000 | 0.1009 | No |
| 108 | ALDH1A1 | 16547 | -0.000 | 0.0812 | No |
| 109 | AR | 16725 | -0.000 | 0.0713 | No |
| 110 | RXRA | 17312 | -0.000 | 0.0386 | No |
| 111 | HSD3B1 | 17597 | -0.000 | 0.0227 | No |