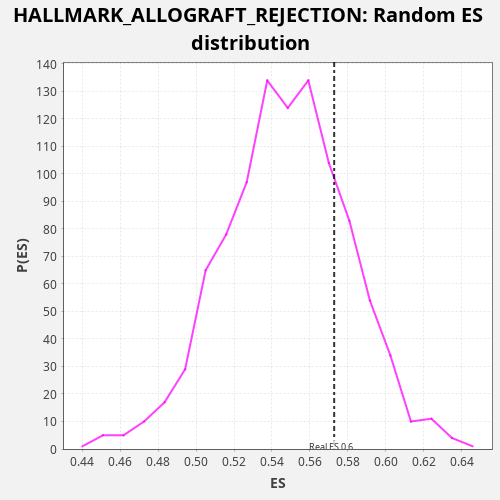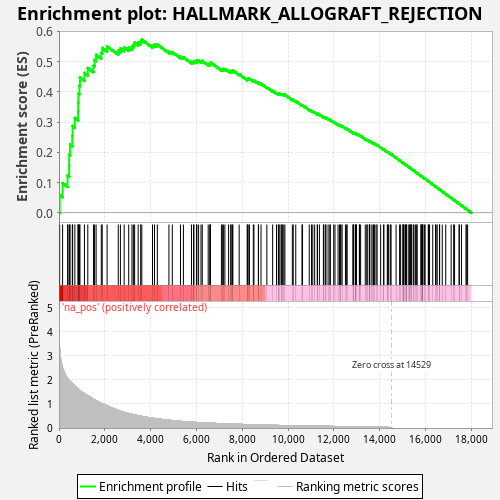
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_ALLOGRAFT_REJECTION |
| Enrichment Score (ES) | 0.57280207 |
| Normalized Enrichment Score (NES) | 1.0459957 |
| Nominal p-value | 0.223 |
| FDR q-value | 0.639952 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | JAK2 | 29 | 3.319 | 0.0592 | Yes |
| 2 | GBP2 | 157 | 2.508 | 0.0981 | Yes |
| 3 | TLR6 | 374 | 2.062 | 0.1237 | Yes |
| 4 | CCND3 | 442 | 1.984 | 0.1563 | Yes |
| 5 | AARS1 | 445 | 1.978 | 0.1925 | Yes |
| 6 | ACVR2A | 484 | 1.935 | 0.2258 | Yes |
| 7 | TPD52 | 581 | 1.850 | 0.2543 | Yes |
| 8 | CD96 | 595 | 1.836 | 0.2873 | Yes |
| 9 | CCR5 | 693 | 1.741 | 0.3137 | Yes |
| 10 | IL2RA | 834 | 1.615 | 0.3355 | Yes |
| 11 | GCNT1 | 839 | 1.611 | 0.3648 | Yes |
| 12 | F2R | 846 | 1.607 | 0.3939 | Yes |
| 13 | THY1 | 891 | 1.571 | 0.4202 | Yes |
| 14 | ACHE | 915 | 1.551 | 0.4473 | Yes |
| 15 | IL18RAP | 1110 | 1.421 | 0.4625 | Yes |
| 16 | GZMB | 1257 | 1.343 | 0.4789 | Yes |
| 17 | TAP2 | 1504 | 1.195 | 0.4870 | Yes |
| 18 | CCL4 | 1553 | 1.169 | 0.5058 | Yes |
| 19 | ITGB2 | 1620 | 1.130 | 0.5228 | Yes |
| 20 | NME1 | 1846 | 1.031 | 0.5290 | Yes |
| 21 | RARS1 | 1887 | 1.012 | 0.5453 | Yes |
| 22 | LY75 | 2100 | 0.923 | 0.5504 | Yes |
| 23 | IRF7 | 2587 | 0.732 | 0.5365 | Yes |
| 24 | SOCS5 | 2681 | 0.699 | 0.5441 | Yes |
| 25 | HCLS1 | 2844 | 0.650 | 0.5469 | Yes |
| 26 | BRCA1 | 3043 | 0.601 | 0.5468 | Yes |
| 27 | HLA-A | 3185 | 0.570 | 0.5493 | Yes |
| 28 | CXCL9 | 3248 | 0.553 | 0.5560 | Yes |
| 29 | DYRK3 | 3302 | 0.542 | 0.5630 | Yes |
| 30 | CD3G | 3456 | 0.513 | 0.5638 | Yes |
| 31 | MTIF2 | 3565 | 0.491 | 0.5667 | Yes |
| 32 | PTPRC | 3615 | 0.482 | 0.5728 | Yes |
| 33 | NCF4 | 4082 | 0.408 | 0.5541 | No |
| 34 | C2 | 4168 | 0.395 | 0.5566 | No |
| 35 | TLR1 | 4292 | 0.379 | 0.5566 | No |
| 36 | CAPG | 4799 | 0.325 | 0.5342 | No |
| 37 | IL16 | 4946 | 0.311 | 0.5317 | No |
| 38 | CD3D | 5304 | 0.279 | 0.5167 | No |
| 39 | IL12RB1 | 5431 | 0.269 | 0.5146 | No |
| 40 | GZMA | 5783 | 0.243 | 0.4993 | No |
| 41 | CRTAM | 5873 | 0.238 | 0.4987 | No |
| 42 | MRPL3 | 5890 | 0.237 | 0.5022 | No |
| 43 | EIF4G3 | 6000 | 0.231 | 0.5003 | No |
| 44 | HLA-DOB | 6021 | 0.230 | 0.5034 | No |
| 45 | IFNAR2 | 6094 | 0.226 | 0.5035 | No |
| 46 | CCL5 | 6199 | 0.221 | 0.5017 | No |
| 47 | LY86 | 6255 | 0.219 | 0.5026 | No |
| 48 | PRF1 | 6513 | 0.205 | 0.4919 | No |
| 49 | STAB1 | 6585 | 0.201 | 0.4916 | No |
| 50 | TLR3 | 6596 | 0.201 | 0.4948 | No |
| 51 | PF4 | 6613 | 0.200 | 0.4975 | No |
| 52 | NCR1 | 7090 | 0.179 | 0.4741 | No |
| 53 | LCK | 7146 | 0.176 | 0.4742 | No |
| 54 | TAP1 | 7176 | 0.175 | 0.4758 | No |
| 55 | ZAP70 | 7241 | 0.173 | 0.4754 | No |
| 56 | ITGAL | 7404 | 0.167 | 0.4693 | No |
| 57 | HDAC9 | 7497 | 0.164 | 0.4672 | No |
| 58 | IL27RA | 7517 | 0.163 | 0.4691 | No |
| 59 | RPL3L | 7578 | 0.161 | 0.4687 | No |
| 60 | PSMB10 | 7588 | 0.161 | 0.4711 | No |
| 61 | LCP2 | 7865 | 0.152 | 0.4584 | No |
| 62 | CD8B | 8214 | 0.140 | 0.4415 | No |
| 63 | IL11 | 8236 | 0.139 | 0.4428 | No |
| 64 | ICAM1 | 8256 | 0.139 | 0.4443 | No |
| 65 | ELANE | 8314 | 0.137 | 0.4436 | No |
| 66 | FYB1 | 8482 | 0.132 | 0.4367 | No |
| 67 | IL12B | 8510 | 0.132 | 0.4376 | No |
| 68 | EREG | 8706 | 0.127 | 0.4290 | No |
| 69 | KLRD1 | 8707 | 0.127 | 0.4313 | No |
| 70 | IFNG | 8819 | 0.124 | 0.4273 | No |
| 71 | CCL11 | 9074 | 0.117 | 0.4152 | No |
| 72 | NLRP3 | 9327 | 0.111 | 0.4031 | No |
| 73 | CD28 | 9495 | 0.107 | 0.3957 | No |
| 74 | EGFR | 9576 | 0.106 | 0.3931 | No |
| 75 | TAPBP | 9592 | 0.105 | 0.3942 | No |
| 76 | CCL22 | 9641 | 0.104 | 0.3934 | No |
| 77 | IFNGR2 | 9703 | 0.103 | 0.3919 | No |
| 78 | CD4 | 9731 | 0.102 | 0.3923 | No |
| 79 | DARS1 | 9765 | 0.101 | 0.3923 | No |
| 80 | CCR2 | 9816 | 0.100 | 0.3913 | No |
| 81 | HIF1A | 9861 | 0.099 | 0.3906 | No |
| 82 | IKBKB | 10186 | 0.094 | 0.3742 | No |
| 83 | FCGR2B | 10226 | 0.093 | 0.3737 | No |
| 84 | F2 | 10336 | 0.091 | 0.3692 | No |
| 85 | BCL10 | 10608 | 0.086 | 0.3556 | No |
| 86 | CXCL13 | 10632 | 0.086 | 0.3559 | No |
| 87 | CD7 | 10925 | 0.081 | 0.3410 | No |
| 88 | RPL9 | 11022 | 0.079 | 0.3370 | No |
| 89 | CCL13 | 11080 | 0.078 | 0.3352 | No |
| 90 | MMP9 | 11161 | 0.077 | 0.3322 | No |
| 91 | IGSF6 | 11277 | 0.075 | 0.3271 | No |
| 92 | CD1D | 11281 | 0.075 | 0.3283 | No |
| 93 | HLA-E | 11368 | 0.074 | 0.3248 | No |
| 94 | KRT1 | 11543 | 0.071 | 0.3163 | No |
| 95 | CD79A | 11559 | 0.071 | 0.3168 | No |
| 96 | ITK | 11628 | 0.069 | 0.3142 | No |
| 97 | IFNGR1 | 11655 | 0.069 | 0.3141 | No |
| 98 | FGR | 11747 | 0.068 | 0.3102 | No |
| 99 | GLMN | 11822 | 0.067 | 0.3073 | No |
| 100 | EIF5A | 11846 | 0.066 | 0.3072 | No |
| 101 | CD8A | 11993 | 0.064 | 0.3001 | No |
| 102 | IRF4 | 12048 | 0.063 | 0.2983 | No |
| 103 | CTSS | 12182 | 0.061 | 0.2919 | No |
| 104 | HLA-DRA | 12248 | 0.061 | 0.2894 | No |
| 105 | PRKCG | 12284 | 0.060 | 0.2885 | No |
| 106 | PTPN6 | 12309 | 0.060 | 0.2883 | No |
| 107 | CD3E | 12361 | 0.059 | 0.2865 | No |
| 108 | IL9 | 12505 | 0.057 | 0.2795 | No |
| 109 | RIPK2 | 12559 | 0.056 | 0.2776 | No |
| 110 | APBB1 | 12577 | 0.056 | 0.2776 | No |
| 111 | NOS2 | 12836 | 0.052 | 0.2641 | No |
| 112 | LTB | 12838 | 0.052 | 0.2650 | No |
| 113 | DEGS1 | 12866 | 0.052 | 0.2645 | No |
| 114 | STAT4 | 12939 | 0.051 | 0.2614 | No |
| 115 | CD40 | 12952 | 0.051 | 0.2616 | No |
| 116 | IL12A | 12958 | 0.051 | 0.2623 | No |
| 117 | HLA-G | 12988 | 0.051 | 0.2616 | No |
| 118 | UBE2D1 | 13112 | 0.049 | 0.2556 | No |
| 119 | ST8SIA4 | 13121 | 0.049 | 0.2560 | No |
| 120 | MBL2 | 13168 | 0.048 | 0.2543 | No |
| 121 | CFP | 13370 | 0.046 | 0.2439 | No |
| 122 | HLA-DOA | 13440 | 0.045 | 0.2408 | No |
| 123 | IL2RB | 13455 | 0.044 | 0.2408 | No |
| 124 | CD74 | 13545 | 0.043 | 0.2366 | No |
| 125 | ELF4 | 13553 | 0.043 | 0.2370 | No |
| 126 | BCAT1 | 13556 | 0.043 | 0.2377 | No |
| 127 | GALNT1 | 13626 | 0.042 | 0.2346 | No |
| 128 | CD40LG | 13704 | 0.041 | 0.2310 | No |
| 129 | IL4R | 13728 | 0.041 | 0.2304 | No |
| 130 | IL15 | 13778 | 0.040 | 0.2284 | No |
| 131 | HLA-DMA | 13841 | 0.039 | 0.2257 | No |
| 132 | ABCE1 | 13887 | 0.038 | 0.2238 | No |
| 133 | CXCR3 | 14042 | 0.035 | 0.2158 | No |
| 134 | WARS1 | 14177 | 0.033 | 0.2089 | No |
| 135 | RPS9 | 14178 | 0.033 | 0.2095 | No |
| 136 | WAS | 14332 | 0.027 | 0.2014 | No |
| 137 | INHBA | 14354 | 0.026 | 0.2007 | No |
| 138 | SPI1 | 14391 | 0.023 | 0.1991 | No |
| 139 | HLA-DQA1 | 14464 | 0.019 | 0.1954 | No |
| 140 | UBE2N | 14497 | 0.016 | 0.1939 | No |
| 141 | BCL3 | 14715 | -0.000 | 0.1817 | No |
| 142 | TGFB2 | 14869 | -0.000 | 0.1731 | No |
| 143 | EIF3D | 14913 | -0.000 | 0.1707 | No |
| 144 | TIMP1 | 15009 | -0.000 | 0.1654 | No |
| 145 | CSK | 15032 | -0.000 | 0.1642 | No |
| 146 | EIF3J | 15035 | -0.000 | 0.1640 | No |
| 147 | IL7 | 15039 | -0.000 | 0.1639 | No |
| 148 | TGFB1 | 15066 | -0.000 | 0.1624 | No |
| 149 | EIF3A | 15129 | -0.000 | 0.1589 | No |
| 150 | CCL7 | 15157 | -0.000 | 0.1574 | No |
| 151 | CCL2 | 15158 | -0.000 | 0.1574 | No |
| 152 | IL2 | 15181 | -0.000 | 0.1562 | No |
| 153 | IL4 | 15275 | -0.000 | 0.1510 | No |
| 154 | CD86 | 15288 | -0.000 | 0.1503 | No |
| 155 | STAT1 | 15318 | -0.000 | 0.1487 | No |
| 156 | CD2 | 15354 | -0.000 | 0.1467 | No |
| 157 | FASLG | 15369 | -0.000 | 0.1459 | No |
| 158 | CCND2 | 15397 | -0.000 | 0.1444 | No |
| 159 | CD80 | 15469 | -0.000 | 0.1404 | No |
| 160 | SRGN | 15497 | -0.000 | 0.1389 | No |
| 161 | IL1B | 15568 | -0.000 | 0.1350 | No |
| 162 | TRAF2 | 15615 | -0.000 | 0.1324 | No |
| 163 | LIF | 15635 | -0.000 | 0.1313 | No |
| 164 | ETS1 | 15799 | -0.000 | 0.1222 | No |
| 165 | MAP3K7 | 15814 | -0.000 | 0.1214 | No |
| 166 | IL6 | 15843 | -0.000 | 0.1198 | No |
| 167 | IL10 | 15855 | -0.000 | 0.1192 | No |
| 168 | ABI1 | 15858 | -0.000 | 0.1191 | No |
| 169 | SIT1 | 15876 | -0.000 | 0.1181 | No |
| 170 | TLR2 | 15888 | -0.000 | 0.1175 | No |
| 171 | GPR65 | 15962 | -0.000 | 0.1134 | No |
| 172 | IRF8 | 15981 | -0.000 | 0.1124 | No |
| 173 | IL2RG | 16121 | -0.000 | 0.1046 | No |
| 174 | CDKN2A | 16138 | -0.000 | 0.1037 | No |
| 175 | IL18 | 16176 | -0.000 | 0.1016 | No |
| 176 | NCK1 | 16316 | -0.000 | 0.0938 | No |
| 177 | INHBB | 16437 | -0.000 | 0.0871 | No |
| 178 | TRAT1 | 16462 | -0.000 | 0.0857 | No |
| 179 | CARTPT | 16511 | -0.000 | 0.0830 | No |
| 180 | PRKCB | 16609 | -0.000 | 0.0776 | No |
| 181 | B2M | 16617 | -0.000 | 0.0772 | No |
| 182 | IL13 | 16732 | -0.000 | 0.0708 | No |
| 183 | CCL19 | 16883 | -0.000 | 0.0624 | No |
| 184 | NPM1 | 17120 | -0.000 | 0.0491 | No |
| 185 | CSF1 | 17230 | -0.000 | 0.0430 | No |
| 186 | SOCS1 | 17268 | -0.000 | 0.0409 | No |
| 187 | CD47 | 17468 | -0.000 | 0.0298 | No |
| 188 | FLNA | 17474 | -0.000 | 0.0295 | No |
| 189 | CD247 | 17570 | -0.000 | 0.0241 | No |
| 190 | TNF | 17767 | -0.000 | 0.0131 | No |
| 191 | HLA-DMB | 17800 | -0.000 | 0.0113 | No |
| 192 | LYN | 17839 | -0.000 | 0.0092 | No |