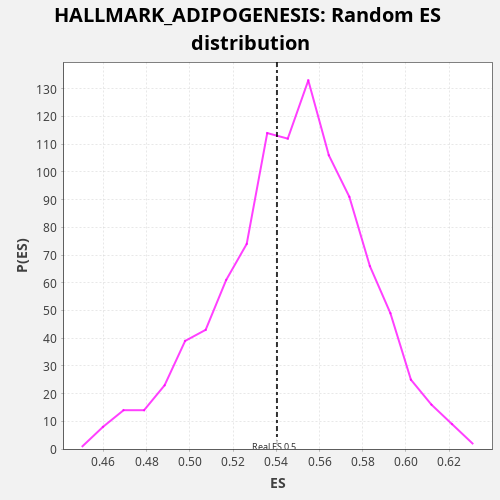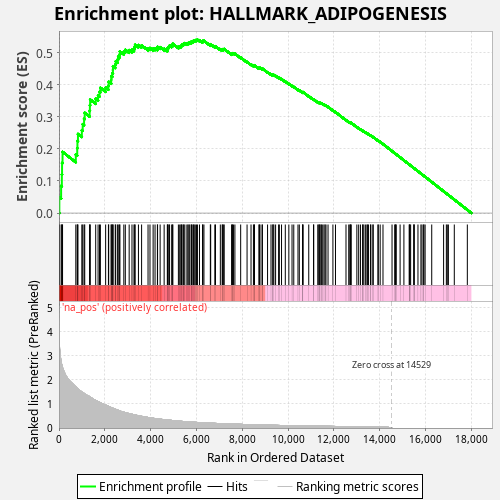
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | HALLMARK_ADIPOGENESIS |
| Enrichment Score (ES) | 0.5402939 |
| Normalized Enrichment Score (NES) | 0.9868542 |
| Nominal p-value | 0.611 |
| FDR q-value | 0.8171102 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | MGLL | 15 | 3.569 | 0.0496 | Yes |
| 2 | STAT5A | 93 | 2.789 | 0.0846 | Yes |
| 3 | COQ5 | 124 | 2.626 | 0.1200 | Yes |
| 4 | PFKFB3 | 134 | 2.603 | 0.1563 | Yes |
| 5 | TANK | 155 | 2.510 | 0.1906 | Yes |
| 6 | DBT | 734 | 1.708 | 0.1823 | Yes |
| 7 | NDUFB7 | 796 | 1.648 | 0.2022 | Yes |
| 8 | BCKDHA | 805 | 1.637 | 0.2248 | Yes |
| 9 | UQCR11 | 828 | 1.620 | 0.2465 | Yes |
| 10 | APLP2 | 997 | 1.498 | 0.2582 | Yes |
| 11 | STOM | 1035 | 1.473 | 0.2769 | Yes |
| 12 | PREB | 1095 | 1.436 | 0.2939 | Yes |
| 13 | SQOR | 1123 | 1.414 | 0.3123 | Yes |
| 14 | PPM1B | 1337 | 1.298 | 0.3187 | Yes |
| 15 | CYC1 | 1349 | 1.288 | 0.3363 | Yes |
| 16 | RIOK3 | 1360 | 1.280 | 0.3538 | Yes |
| 17 | QDPR | 1602 | 1.142 | 0.3564 | Yes |
| 18 | UCK1 | 1706 | 1.096 | 0.3661 | Yes |
| 19 | ITGA7 | 1769 | 1.068 | 0.3777 | Yes |
| 20 | PPP1R15B | 1806 | 1.051 | 0.3905 | Yes |
| 21 | NDUFA5 | 2036 | 0.949 | 0.3911 | Yes |
| 22 | RTN3 | 2158 | 0.897 | 0.3970 | Yes |
| 23 | DLD | 2169 | 0.894 | 0.4090 | Yes |
| 24 | CIDEA | 2279 | 0.847 | 0.4149 | Yes |
| 25 | MAP4K3 | 2282 | 0.846 | 0.4267 | Yes |
| 26 | GBE1 | 2328 | 0.829 | 0.4359 | Yes |
| 27 | CRAT | 2354 | 0.819 | 0.4461 | Yes |
| 28 | PGM1 | 2357 | 0.818 | 0.4575 | Yes |
| 29 | BCL2L13 | 2456 | 0.780 | 0.4630 | Yes |
| 30 | CHUK | 2476 | 0.773 | 0.4729 | Yes |
| 31 | DLAT | 2560 | 0.743 | 0.4787 | Yes |
| 32 | UQCRC1 | 2584 | 0.733 | 0.4878 | Yes |
| 33 | HIBCH | 2647 | 0.711 | 0.4943 | Yes |
| 34 | ACLY | 2656 | 0.706 | 0.5038 | Yes |
| 35 | PFKL | 2830 | 0.654 | 0.5034 | Yes |
| 36 | CMBL | 2895 | 0.638 | 0.5088 | Yes |
| 37 | PRDX3 | 3063 | 0.596 | 0.5078 | Yes |
| 38 | SUCLG1 | 3186 | 0.570 | 0.5090 | Yes |
| 39 | TALDO1 | 3273 | 0.548 | 0.5119 | Yes |
| 40 | NDUFS3 | 3303 | 0.542 | 0.5179 | Yes |
| 41 | CAT | 3323 | 0.539 | 0.5245 | Yes |
| 42 | DHCR7 | 3468 | 0.510 | 0.5236 | Yes |
| 43 | MCCC1 | 3606 | 0.484 | 0.5228 | Yes |
| 44 | MDH2 | 3887 | 0.435 | 0.5132 | Yes |
| 45 | PHLDB1 | 3972 | 0.423 | 0.5144 | Yes |
| 46 | DHRS7B | 4109 | 0.404 | 0.5125 | Yes |
| 47 | GHITM | 4193 | 0.391 | 0.5134 | Yes |
| 48 | GPX3 | 4296 | 0.379 | 0.5130 | Yes |
| 49 | PHYH | 4303 | 0.377 | 0.5180 | Yes |
| 50 | SULT1A1 | 4415 | 0.365 | 0.5169 | Yes |
| 51 | SPARCL1 | 4590 | 0.346 | 0.5120 | Yes |
| 52 | ADCY6 | 4732 | 0.331 | 0.5088 | Yes |
| 53 | CD302 | 4733 | 0.331 | 0.5135 | Yes |
| 54 | MYLK | 4760 | 0.329 | 0.5166 | Yes |
| 55 | NABP1 | 4801 | 0.325 | 0.5190 | Yes |
| 56 | LTC4S | 4827 | 0.322 | 0.5221 | Yes |
| 57 | ABCA1 | 4919 | 0.313 | 0.5214 | Yes |
| 58 | DHRS7 | 4961 | 0.309 | 0.5235 | Yes |
| 59 | NKIRAS1 | 4967 | 0.309 | 0.5276 | Yes |
| 60 | ACADS | 5212 | 0.287 | 0.5179 | Yes |
| 61 | SCARB1 | 5270 | 0.281 | 0.5187 | Yes |
| 62 | SLC27A1 | 5319 | 0.278 | 0.5200 | Yes |
| 63 | ACADM | 5346 | 0.275 | 0.5224 | Yes |
| 64 | TST | 5370 | 0.273 | 0.5249 | Yes |
| 65 | SLC25A1 | 5425 | 0.269 | 0.5257 | Yes |
| 66 | ACAA2 | 5448 | 0.267 | 0.5282 | Yes |
| 67 | ABCB8 | 5493 | 0.264 | 0.5295 | Yes |
| 68 | MRPL15 | 5584 | 0.258 | 0.5281 | Yes |
| 69 | COX6A1 | 5627 | 0.254 | 0.5293 | Yes |
| 70 | SLC66A3 | 5670 | 0.251 | 0.5305 | Yes |
| 71 | LIPE | 5708 | 0.248 | 0.5319 | Yes |
| 72 | PDCD4 | 5775 | 0.244 | 0.5317 | Yes |
| 73 | EPHX2 | 5781 | 0.244 | 0.5348 | Yes |
| 74 | DNAJC15 | 5844 | 0.240 | 0.5347 | Yes |
| 75 | SLC19A1 | 5867 | 0.238 | 0.5369 | Yes |
| 76 | DECR1 | 5903 | 0.236 | 0.5382 | Yes |
| 77 | COQ9 | 5949 | 0.234 | 0.5390 | Yes |
| 78 | PLIN2 | 6004 | 0.231 | 0.5392 | Yes |
| 79 | FAH | 6044 | 0.229 | 0.5403 | Yes |
| 80 | ALDH2 | 6132 | 0.224 | 0.5386 | No |
| 81 | FABP4 | 6266 | 0.218 | 0.5342 | No |
| 82 | ITIH5 | 6280 | 0.217 | 0.5365 | No |
| 83 | OMD | 6327 | 0.214 | 0.5370 | No |
| 84 | GPAM | 6607 | 0.200 | 0.5241 | No |
| 85 | CD36 | 6622 | 0.199 | 0.5261 | No |
| 86 | IDH1 | 6803 | 0.191 | 0.5187 | No |
| 87 | COQ3 | 6832 | 0.189 | 0.5198 | No |
| 88 | RETSAT | 7043 | 0.181 | 0.5106 | No |
| 89 | RMDN3 | 7127 | 0.177 | 0.5084 | No |
| 90 | RAB34 | 7133 | 0.177 | 0.5107 | No |
| 91 | ECHS1 | 7188 | 0.175 | 0.5101 | No |
| 92 | CMPK1 | 7210 | 0.174 | 0.5114 | No |
| 93 | SAMM50 | 7536 | 0.163 | 0.4954 | No |
| 94 | LPL | 7560 | 0.162 | 0.4964 | No |
| 95 | SLC25A10 | 7571 | 0.161 | 0.4981 | No |
| 96 | SSPN | 7616 | 0.160 | 0.4979 | No |
| 97 | AK2 | 7679 | 0.158 | 0.4967 | No |
| 98 | TKT | 7931 | 0.150 | 0.4847 | No |
| 99 | CPT2 | 8210 | 0.140 | 0.4711 | No |
| 100 | ACADL | 8383 | 0.135 | 0.4633 | No |
| 101 | UCP2 | 8488 | 0.132 | 0.4593 | No |
| 102 | UBC | 8519 | 0.132 | 0.4595 | No |
| 103 | ESYT1 | 8534 | 0.131 | 0.4606 | No |
| 104 | ACOX1 | 8727 | 0.126 | 0.4516 | No |
| 105 | ATL2 | 8749 | 0.126 | 0.4522 | No |
| 106 | ARAF | 8774 | 0.125 | 0.4526 | No |
| 107 | ATP5PO | 8864 | 0.123 | 0.4493 | No |
| 108 | ITSN1 | 8880 | 0.122 | 0.4502 | No |
| 109 | SCP2 | 9105 | 0.117 | 0.4393 | No |
| 110 | GPX4 | 9255 | 0.112 | 0.4325 | No |
| 111 | MIGA2 | 9337 | 0.111 | 0.4295 | No |
| 112 | MRAP | 9339 | 0.110 | 0.4310 | No |
| 113 | IMMT | 9380 | 0.109 | 0.4303 | No |
| 114 | UQCR10 | 9456 | 0.108 | 0.4276 | No |
| 115 | SORBS1 | 9587 | 0.105 | 0.4218 | No |
| 116 | MTCH2 | 9619 | 0.105 | 0.4216 | No |
| 117 | SLC5A6 | 9715 | 0.103 | 0.4177 | No |
| 118 | CS | 9879 | 0.099 | 0.4099 | No |
| 119 | ELMOD3 | 10034 | 0.096 | 0.4027 | No |
| 120 | AGPAT3 | 10173 | 0.094 | 0.3962 | No |
| 121 | CCNG2 | 10249 | 0.092 | 0.3933 | No |
| 122 | FZD4 | 10433 | 0.089 | 0.3843 | No |
| 123 | ADIPOQ | 10491 | 0.088 | 0.3824 | No |
| 124 | CD151 | 10630 | 0.086 | 0.3758 | No |
| 125 | POR | 10648 | 0.086 | 0.3761 | No |
| 126 | ENPP2 | 10650 | 0.086 | 0.3772 | No |
| 127 | IDH3A | 10903 | 0.081 | 0.3642 | No |
| 128 | SOWAHC | 11112 | 0.077 | 0.3536 | No |
| 129 | SDHB | 11120 | 0.077 | 0.3543 | No |
| 130 | MGST3 | 11297 | 0.075 | 0.3455 | No |
| 131 | ALDOA | 11342 | 0.074 | 0.3441 | No |
| 132 | REEP5 | 11362 | 0.074 | 0.3441 | No |
| 133 | ADIG | 11387 | 0.073 | 0.3437 | No |
| 134 | CAVIN1 | 11438 | 0.073 | 0.3420 | No |
| 135 | SDHC | 11452 | 0.072 | 0.3423 | No |
| 136 | VEGFB | 11513 | 0.071 | 0.3399 | No |
| 137 | COL4A1 | 11569 | 0.071 | 0.3378 | No |
| 138 | ETFB | 11616 | 0.070 | 0.3362 | No |
| 139 | IFNGR1 | 11655 | 0.069 | 0.3350 | No |
| 140 | SOD1 | 11745 | 0.068 | 0.3310 | No |
| 141 | ACO2 | 11958 | 0.064 | 0.3200 | No |
| 142 | CYP4B1 | 12070 | 0.063 | 0.3147 | No |
| 143 | COL15A1 | 12529 | 0.057 | 0.2898 | No |
| 144 | JAGN1 | 12649 | 0.055 | 0.2839 | No |
| 145 | HSPB8 | 12697 | 0.054 | 0.2820 | No |
| 146 | LPCAT3 | 12720 | 0.054 | 0.2815 | No |
| 147 | BAZ2A | 12734 | 0.054 | 0.2815 | No |
| 148 | CAVIN2 | 12756 | 0.053 | 0.2811 | No |
| 149 | DNAJB9 | 13003 | 0.050 | 0.2680 | No |
| 150 | PTCD3 | 13083 | 0.049 | 0.2643 | No |
| 151 | MTARC2 | 13160 | 0.048 | 0.2607 | No |
| 152 | ORM1 | 13257 | 0.047 | 0.2560 | No |
| 153 | SLC1A5 | 13265 | 0.047 | 0.2562 | No |
| 154 | RREB1 | 13311 | 0.046 | 0.2544 | No |
| 155 | ADIPOR2 | 13387 | 0.045 | 0.2508 | No |
| 156 | LIFR | 13445 | 0.044 | 0.2482 | No |
| 157 | UQCRQ | 13474 | 0.044 | 0.2473 | No |
| 158 | ANGPT1 | 13509 | 0.044 | 0.2460 | No |
| 159 | GPAT4 | 13597 | 0.042 | 0.2417 | No |
| 160 | GRPEL1 | 13615 | 0.042 | 0.2413 | No |
| 161 | LAMA4 | 13703 | 0.041 | 0.2370 | No |
| 162 | PEX14 | 13716 | 0.041 | 0.2369 | No |
| 163 | C3 | 13925 | 0.037 | 0.2258 | No |
| 164 | PPARG | 13937 | 0.037 | 0.2257 | No |
| 165 | ESRRA | 14021 | 0.036 | 0.2215 | No |
| 166 | PIM3 | 14144 | 0.033 | 0.2151 | No |
| 167 | NDUFAB1 | 14539 | -0.000 | 0.1930 | No |
| 168 | PTGER3 | 14656 | -0.000 | 0.1865 | No |
| 169 | ME1 | 14685 | -0.000 | 0.1849 | No |
| 170 | IDH3G | 14701 | -0.000 | 0.1841 | No |
| 171 | ATP1B3 | 14721 | -0.000 | 0.1830 | No |
| 172 | DDT | 14891 | -0.000 | 0.1735 | No |
| 173 | RETN | 15057 | -0.000 | 0.1643 | No |
| 174 | BCL6 | 15287 | -0.000 | 0.1514 | No |
| 175 | GPD2 | 15307 | -0.000 | 0.1504 | No |
| 176 | REEP6 | 15312 | -0.000 | 0.1501 | No |
| 177 | GADD45A | 15351 | -0.000 | 0.1480 | No |
| 178 | ARL4A | 15491 | -0.000 | 0.1402 | No |
| 179 | CDKN2C | 15501 | -0.000 | 0.1397 | No |
| 180 | RNF11 | 15502 | -0.000 | 0.1397 | No |
| 181 | APOE | 15671 | -0.000 | 0.1303 | No |
| 182 | UBQLN1 | 15800 | -0.000 | 0.1231 | No |
| 183 | NMT1 | 15848 | -0.000 | 0.1204 | No |
| 184 | G3BP2 | 15921 | -0.000 | 0.1164 | No |
| 185 | HADH | 15926 | -0.000 | 0.1162 | No |
| 186 | TOB1 | 15983 | -0.000 | 0.1130 | No |
| 187 | DRAM2 | 16272 | -0.000 | 0.0969 | No |
| 188 | ELOVL6 | 16790 | -0.000 | 0.0678 | No |
| 189 | SNCG | 16910 | -0.000 | 0.0611 | No |
| 190 | LEP | 16950 | -0.000 | 0.0590 | No |
| 191 | COX8A | 16999 | -0.000 | 0.0563 | No |
| 192 | DGAT1 | 17257 | -0.000 | 0.0418 | No |
| 193 | CHCHD10 | 17826 | -0.000 | 0.0099 | No |