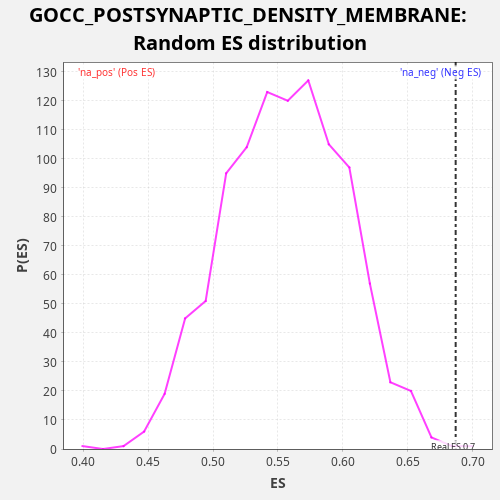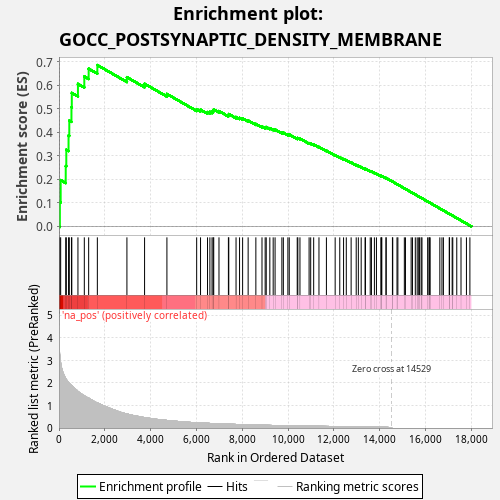
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | GOCC_POSTSYNAPTIC_DENSITY_MEMBRANE |
| Enrichment Score (ES) | 0.6868035 |
| Normalized Enrichment Score (NES) | 1.2346699 |
| Nominal p-value | 0.001 |
| FDR q-value | 0.48326123 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | CACNG5 | 38 | 3.206 | 0.1035 | Yes |
| 2 | DAGLA | 73 | 2.896 | 0.1971 | Yes |
| 3 | EFNB3 | 294 | 2.187 | 0.2568 | Yes |
| 4 | RGS7BP | 319 | 2.151 | 0.3264 | Yes |
| 5 | SLC16A7 | 417 | 2.010 | 0.3872 | Yes |
| 6 | SORCS3 | 450 | 1.973 | 0.4504 | Yes |
| 7 | GRIK4 | 544 | 1.886 | 0.5074 | Yes |
| 8 | DCC | 559 | 1.871 | 0.5683 | Yes |
| 9 | NETO1 | 827 | 1.621 | 0.6068 | Yes |
| 10 | RGS9 | 1105 | 1.428 | 0.6383 | Yes |
| 11 | DLG2 | 1295 | 1.323 | 0.6714 | Yes |
| 12 | RNF10 | 1674 | 1.108 | 0.6868 | Yes |
| 13 | SEMA4F | 2965 | 0.620 | 0.6352 | No |
| 14 | LIN7B | 3736 | 0.459 | 0.6073 | No |
| 15 | NRCAM | 4711 | 0.333 | 0.5639 | No |
| 16 | PRRT2 | 6010 | 0.231 | 0.4990 | No |
| 17 | SORCS2 | 6179 | 0.222 | 0.4969 | No |
| 18 | SEMA4B | 6484 | 0.206 | 0.4868 | No |
| 19 | ITGA8 | 6587 | 0.201 | 0.4877 | No |
| 20 | GRIN2C | 6681 | 0.196 | 0.4890 | No |
| 21 | SIGMAR1 | 6731 | 0.194 | 0.4926 | No |
| 22 | GRIK1 | 6755 | 0.193 | 0.4977 | No |
| 23 | CACNG8 | 6984 | 0.183 | 0.4910 | No |
| 24 | CACNA1C | 7395 | 0.167 | 0.4736 | No |
| 25 | PTPRS | 7412 | 0.167 | 0.4782 | No |
| 26 | OPRD1 | 7729 | 0.156 | 0.4657 | No |
| 27 | GRIN3A | 7882 | 0.152 | 0.4622 | No |
| 28 | ATP2B2 | 8016 | 0.147 | 0.4596 | No |
| 29 | GRIN3B | 8257 | 0.139 | 0.4508 | No |
| 30 | PTPRO | 8594 | 0.129 | 0.4363 | No |
| 31 | KCNH1 | 8862 | 0.123 | 0.4254 | No |
| 32 | GRIA1 | 9000 | 0.119 | 0.4217 | No |
| 33 | GRIA4 | 9047 | 0.118 | 0.4230 | No |
| 34 | TIAM1 | 9204 | 0.114 | 0.4181 | No |
| 35 | LRFN3 | 9350 | 0.110 | 0.4136 | No |
| 36 | SYNDIG1 | 9433 | 0.108 | 0.4126 | No |
| 37 | NETO2 | 9730 | 0.102 | 0.3995 | No |
| 38 | GRM1 | 9794 | 0.101 | 0.3993 | No |
| 39 | AKAP9 | 9992 | 0.097 | 0.3915 | No |
| 40 | LRFN5 | 10052 | 0.096 | 0.3913 | No |
| 41 | DLG1 | 10400 | 0.090 | 0.3749 | No |
| 42 | GRIN2D | 10426 | 0.090 | 0.3765 | No |
| 43 | LRFN4 | 10519 | 0.088 | 0.3742 | No |
| 44 | CLSTN3 | 10918 | 0.081 | 0.3547 | No |
| 45 | ACTN2 | 10988 | 0.079 | 0.3534 | No |
| 46 | ERBB4 | 11118 | 0.077 | 0.3488 | No |
| 47 | SEMA4C | 11349 | 0.074 | 0.3384 | No |
| 48 | SHISA6 | 11674 | 0.069 | 0.3225 | No |
| 49 | LRFN2 | 12056 | 0.063 | 0.3033 | No |
| 50 | ASIC1 | 12257 | 0.060 | 0.2942 | No |
| 51 | ADRA2A | 12423 | 0.058 | 0.2869 | No |
| 52 | LIN7C | 12542 | 0.056 | 0.2821 | No |
| 53 | PTPRT | 12753 | 0.054 | 0.2722 | No |
| 54 | SLITRK3 | 12975 | 0.051 | 0.2615 | No |
| 55 | LIN7A | 13068 | 0.049 | 0.2580 | No |
| 56 | PRRT1 | 13192 | 0.048 | 0.2527 | No |
| 57 | EPHA4 | 13363 | 0.046 | 0.2447 | No |
| 58 | GSG1L | 13378 | 0.045 | 0.2454 | No |
| 59 | ABHD17A | 13587 | 0.042 | 0.2352 | No |
| 60 | GRIK2 | 13640 | 0.042 | 0.2337 | No |
| 61 | SCN8A | 13769 | 0.040 | 0.2278 | No |
| 62 | ADCY1 | 13858 | 0.039 | 0.2242 | No |
| 63 | GRIK3 | 14052 | 0.035 | 0.2146 | No |
| 64 | LRRTM3 | 14076 | 0.035 | 0.2144 | No |
| 65 | LRFN1 | 14093 | 0.034 | 0.2147 | No |
| 66 | LRRC4B | 14276 | 0.029 | 0.2055 | No |
| 67 | NLGN4X | 14282 | 0.029 | 0.2062 | No |
| 68 | CACNG3 | 14556 | -0.000 | 0.1909 | No |
| 69 | ADAM22 | 14570 | -0.000 | 0.1902 | No |
| 70 | CACNG4 | 14756 | -0.000 | 0.1799 | No |
| 71 | DLG3 | 14794 | -0.000 | 0.1778 | No |
| 72 | CACNG7 | 15076 | -0.000 | 0.1621 | No |
| 73 | ABHD17B | 15125 | -0.000 | 0.1594 | No |
| 74 | PLPPR4 | 15373 | -0.000 | 0.1456 | No |
| 75 | GRIA2 | 15431 | -0.000 | 0.1425 | No |
| 76 | EFNB2 | 15555 | -0.000 | 0.1356 | No |
| 77 | LRRC4 | 15637 | -0.000 | 0.1311 | No |
| 78 | PRR7 | 15704 | -0.000 | 0.1274 | No |
| 79 | DLG4 | 15742 | -0.000 | 0.1253 | No |
| 80 | ADGRB3 | 15811 | -0.000 | 0.1215 | No |
| 81 | ABHD17C | 15846 | -0.000 | 0.1196 | No |
| 82 | LRRTM2 | 16094 | -0.000 | 0.1058 | No |
| 83 | LRRC4C | 16154 | -0.000 | 0.1025 | No |
| 84 | CNKSR2 | 16169 | -0.000 | 0.1017 | No |
| 85 | GRID2 | 16203 | -0.000 | 0.0999 | No |
| 86 | CACNG2 | 16626 | -0.000 | 0.0763 | No |
| 87 | GRM5 | 16718 | -0.000 | 0.0713 | No |
| 88 | SLC30A1 | 16782 | -0.000 | 0.0677 | No |
| 89 | NECTIN3 | 17036 | -0.000 | 0.0536 | No |
| 90 | SLITRK1 | 17049 | -0.000 | 0.0529 | No |
| 91 | GRID1 | 17164 | -0.000 | 0.0466 | No |
| 92 | GRIN2A | 17191 | -0.000 | 0.0451 | No |
| 93 | SHISA7 | 17365 | -0.000 | 0.0355 | No |
| 94 | ARC | 17550 | -0.000 | 0.0252 | No |
| 95 | SHISA9 | 17787 | -0.000 | 0.0120 | No |
| 96 | GRIN2B | 17938 | -0.000 | 0.0036 | No |