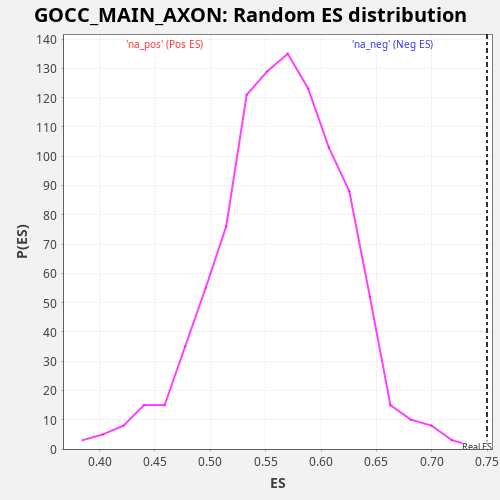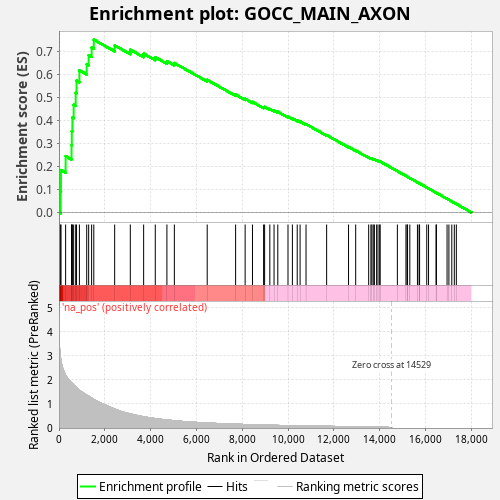
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | GOCC_MAIN_AXON |
| Enrichment Score (ES) | 0.7499938 |
| Normalized Enrichment Score (NES) | 1.3297808 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.33105728 |
| FWER p-Value | 0.866 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | DAGLA | 73 | 2.896 | 0.0910 | Yes |
| 2 | UCN3 | 84 | 2.832 | 0.1834 | Yes |
| 3 | SCN1B | 289 | 2.196 | 0.2442 | Yes |
| 4 | SPTBN4 | 547 | 1.881 | 0.2916 | Yes |
| 5 | GABBR1 | 558 | 1.871 | 0.3525 | Yes |
| 6 | ANK3 | 585 | 1.847 | 0.4117 | Yes |
| 7 | NFASC | 640 | 1.788 | 0.4674 | Yes |
| 8 | MAPT | 729 | 1.712 | 0.5187 | Yes |
| 9 | KCNQ2 | 766 | 1.684 | 0.5720 | Yes |
| 10 | THY1 | 891 | 1.571 | 0.6167 | Yes |
| 11 | CNTNAP1 | 1210 | 1.363 | 0.6437 | Yes |
| 12 | DLG2 | 1295 | 1.323 | 0.6825 | Yes |
| 13 | MAP1A | 1424 | 1.248 | 0.7163 | Yes |
| 14 | NCMAP | 1519 | 1.185 | 0.7500 | Yes |
| 15 | PARD3 | 2430 | 0.790 | 0.7252 | No |
| 16 | MYOC | 3113 | 0.585 | 0.7064 | No |
| 17 | LGI3 | 3695 | 0.466 | 0.6893 | No |
| 18 | CNGA3 | 4203 | 0.390 | 0.6739 | No |
| 19 | NRCAM | 4711 | 0.333 | 0.6566 | No |
| 20 | CNTN2 | 5036 | 0.302 | 0.6484 | No |
| 21 | HAPLN2 | 6470 | 0.207 | 0.5753 | No |
| 22 | EPB41L3 | 7710 | 0.157 | 0.5114 | No |
| 23 | BIN1 | 8126 | 0.143 | 0.4930 | No |
| 24 | IQCJ-SCHIP1 | 8446 | 0.133 | 0.4796 | No |
| 25 | ANK1 | 8932 | 0.121 | 0.4565 | No |
| 26 | KCNAB2 | 8976 | 0.120 | 0.4581 | No |
| 27 | TIAM1 | 9204 | 0.114 | 0.4491 | No |
| 28 | ERMN | 9388 | 0.109 | 0.4425 | No |
| 29 | KCNC2 | 9552 | 0.106 | 0.4369 | No |
| 30 | MAG | 9997 | 0.097 | 0.4154 | No |
| 31 | CRHBP | 10189 | 0.094 | 0.4078 | No |
| 32 | DLG1 | 10400 | 0.090 | 0.3990 | No |
| 33 | KCNAB1 | 10525 | 0.088 | 0.3950 | No |
| 34 | KCNQ3 | 10784 | 0.083 | 0.3834 | No |
| 35 | C9orf72 | 11684 | 0.068 | 0.3355 | No |
| 36 | TNFRSF1B | 12640 | 0.055 | 0.2841 | No |
| 37 | CD40 | 12952 | 0.051 | 0.2684 | No |
| 38 | DAG1 | 13519 | 0.043 | 0.2383 | No |
| 39 | SCN2A | 13617 | 0.042 | 0.2343 | No |
| 40 | SPOCK1 | 13660 | 0.041 | 0.2333 | No |
| 41 | TUBB4A | 13719 | 0.041 | 0.2314 | No |
| 42 | SCN8A | 13769 | 0.040 | 0.2300 | No |
| 43 | ADORA2A | 13866 | 0.039 | 0.2259 | No |
| 44 | TRIM46 | 13926 | 0.037 | 0.2238 | No |
| 45 | MYO1D | 14001 | 0.036 | 0.2209 | No |
| 46 | MBP | 14018 | 0.036 | 0.2212 | No |
| 47 | MAP2 | 14772 | -0.000 | 0.1792 | No |
| 48 | LGI1 | 15147 | -0.000 | 0.1584 | No |
| 49 | SLC1A2 | 15196 | -0.000 | 0.1557 | No |
| 50 | KCNA1 | 15216 | -0.000 | 0.1546 | No |
| 51 | SPTBN1 | 15315 | -0.000 | 0.1492 | No |
| 52 | KCNC1 | 15648 | -0.000 | 0.1307 | No |
| 53 | MAP1B | 15719 | -0.000 | 0.1267 | No |
| 54 | DLG4 | 15742 | -0.000 | 0.1255 | No |
| 55 | SCN1A | 16054 | -0.000 | 0.1082 | No |
| 56 | CRH | 16133 | -0.000 | 0.1038 | No |
| 57 | ADORA1 | 16460 | -0.000 | 0.0857 | No |
| 58 | UCN | 16477 | -0.000 | 0.0848 | No |
| 59 | CNTNAP2 | 16941 | -0.000 | 0.0590 | No |
| 60 | KCNA2 | 17022 | -0.000 | 0.0545 | No |
| 61 | KCNA4 | 17148 | -0.000 | 0.0475 | No |
| 62 | ROBO2 | 17258 | -0.000 | 0.0415 | No |
| 63 | KCNJ11 | 17346 | -0.000 | 0.0366 | No |