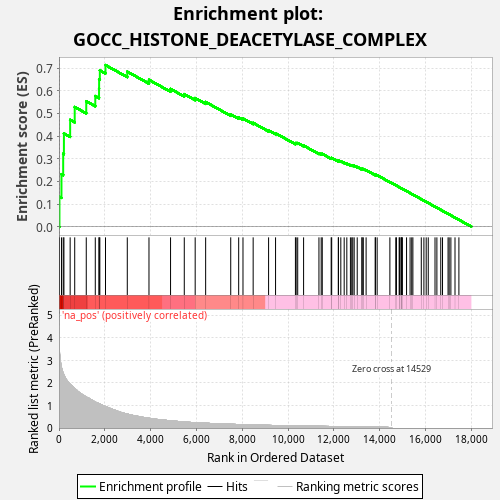
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | GOCC_HISTONE_DEACETYLASE_COMPLEX |
| Enrichment Score (ES) | 0.7134255 |
| Normalized Enrichment Score (NES) | 1.2765365 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.4252737 |
| FWER p-Value | 0.995 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | DMAP1 | 20 | 3.428 | 0.1320 | Yes |
| 2 | CBX5 | 108 | 2.688 | 0.2316 | Yes |
| 3 | GATAD2A | 183 | 2.432 | 0.3219 | Yes |
| 4 | ING3 | 215 | 2.355 | 0.4116 | Yes |
| 5 | MIDEAS | 487 | 1.933 | 0.4715 | Yes |
| 6 | ING2 | 686 | 1.746 | 0.5283 | Yes |
| 7 | SAP130 | 1190 | 1.375 | 0.5537 | Yes |
| 8 | SETD5 | 1584 | 1.155 | 0.5766 | Yes |
| 9 | RERE | 1748 | 1.075 | 0.6093 | Yes |
| 10 | HDAC10 | 1752 | 1.074 | 0.6508 | Yes |
| 11 | CHD5 | 1783 | 1.059 | 0.6902 | Yes |
| 12 | CHD4 | 2030 | 0.951 | 0.7134 | Yes |
| 13 | ZNF541 | 2981 | 0.617 | 0.6844 | No |
| 14 | ING1 | 3928 | 0.429 | 0.6483 | No |
| 15 | BRD8 | 4870 | 0.318 | 0.6082 | No |
| 16 | ARID4A | 5469 | 0.266 | 0.5851 | No |
| 17 | HDAC5 | 5945 | 0.234 | 0.5677 | No |
| 18 | SAP18 | 6402 | 0.210 | 0.5504 | No |
| 19 | HDAC9 | 7497 | 0.164 | 0.4958 | No |
| 20 | ACTR6 | 7844 | 0.153 | 0.4824 | No |
| 21 | HR | 8032 | 0.146 | 0.4777 | No |
| 22 | BRMS1 | 8476 | 0.133 | 0.4581 | No |
| 23 | MBD2 | 9150 | 0.115 | 0.4250 | No |
| 24 | HDAC11 | 9454 | 0.108 | 0.4123 | No |
| 25 | SIN3B | 10321 | 0.091 | 0.3676 | No |
| 26 | NCOR1 | 10352 | 0.091 | 0.3694 | No |
| 27 | CFDP1 | 10421 | 0.090 | 0.3691 | No |
| 28 | EP400 | 10678 | 0.085 | 0.3581 | No |
| 29 | TET1 | 11348 | 0.074 | 0.3237 | No |
| 30 | TRRAP | 11446 | 0.072 | 0.3211 | No |
| 31 | KMT2E | 11493 | 0.072 | 0.3213 | No |
| 32 | MTA3 | 11882 | 0.066 | 0.3022 | No |
| 33 | MTA1 | 11905 | 0.065 | 0.3035 | No |
| 34 | DNTTIP1 | 12194 | 0.061 | 0.2898 | No |
| 35 | MECOM | 12200 | 0.061 | 0.2919 | No |
| 36 | CHD3 | 12303 | 0.060 | 0.2886 | No |
| 37 | RCOR2 | 12450 | 0.058 | 0.2827 | No |
| 38 | ZNF217 | 12565 | 0.056 | 0.2785 | No |
| 39 | HDAC1 | 12722 | 0.054 | 0.2719 | No |
| 40 | CDK2AP1 | 12781 | 0.053 | 0.2707 | No |
| 41 | KAT5 | 12826 | 0.053 | 0.2703 | No |
| 42 | SRCAP | 12886 | 0.052 | 0.2690 | No |
| 43 | HINT1 | 13019 | 0.050 | 0.2636 | No |
| 44 | RCOR3 | 13223 | 0.047 | 0.2541 | No |
| 45 | TRERF1 | 13234 | 0.047 | 0.2554 | No |
| 46 | HDAC3 | 13285 | 0.047 | 0.2544 | No |
| 47 | JMJD1C | 13407 | 0.045 | 0.2494 | No |
| 48 | ARID4B | 13808 | 0.039 | 0.2286 | No |
| 49 | SUDS3 | 13810 | 0.039 | 0.2301 | No |
| 50 | SAP30L | 13889 | 0.038 | 0.2272 | No |
| 51 | RUVBL2 | 14445 | 0.020 | 0.1971 | No |
| 52 | HDAC4 | 14703 | -0.000 | 0.1827 | No |
| 53 | MBD3 | 14732 | -0.000 | 0.1812 | No |
| 54 | RCOR1 | 14850 | -0.000 | 0.1746 | No |
| 55 | HDAC6 | 14870 | -0.000 | 0.1736 | No |
| 56 | BRMS1L | 14931 | -0.000 | 0.1702 | No |
| 57 | CSNK2A1 | 14948 | -0.000 | 0.1693 | No |
| 58 | TBL1X | 14970 | -0.000 | 0.1682 | No |
| 59 | RBBP7 | 14989 | -0.000 | 0.1672 | No |
| 60 | PHF12 | 15173 | -0.000 | 0.1570 | No |
| 61 | KDM3A | 15327 | -0.000 | 0.1484 | No |
| 62 | SATB2 | 15399 | -0.000 | 0.1445 | No |
| 63 | KDM3B | 15449 | -0.000 | 0.1417 | No |
| 64 | PHF21A | 15816 | -0.000 | 0.1213 | No |
| 65 | SINHCAF | 15930 | -0.000 | 0.1150 | No |
| 66 | ANP32E | 16030 | -0.000 | 0.1095 | No |
| 67 | OGT | 16120 | -0.000 | 0.1045 | No |
| 68 | RBBP4 | 16415 | -0.000 | 0.0881 | No |
| 69 | SAP30 | 16497 | -0.000 | 0.0836 | No |
| 70 | CDK2AP2 | 16666 | -0.000 | 0.0742 | No |
| 71 | SIN3A | 16743 | -0.000 | 0.0700 | No |
| 72 | RUVBL1 | 16985 | -0.000 | 0.0566 | No |
| 73 | TBL1XR1 | 17033 | -0.000 | 0.0539 | No |
| 74 | NRIP1 | 17108 | -0.000 | 0.0498 | No |
| 75 | MORF4L1 | 17288 | -0.000 | 0.0398 | No |
| 76 | HDAC2 | 17458 | -0.000 | 0.0304 | No |