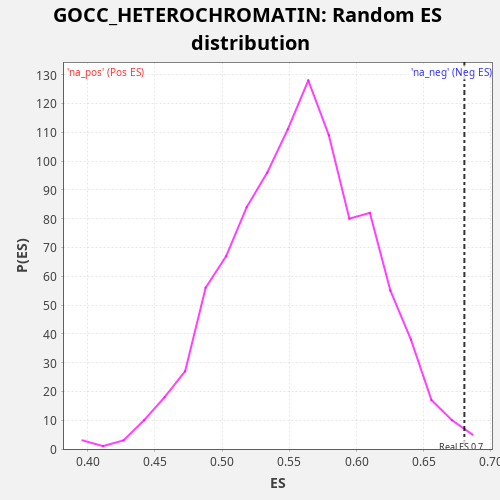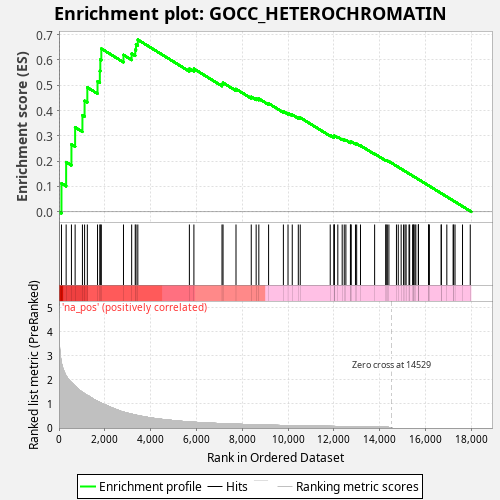
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Chronic fatigue syndrome__20002-both_sexes-1482 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | GOCC_HETEROCHROMATIN |
| Enrichment Score (ES) | 0.6802368 |
| Normalized Enrichment Score (NES) | 1.2193221 |
| Nominal p-value | 0.004 |
| FDR q-value | 0.50505984 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | CBX5 | 108 | 2.688 | 0.1124 | Yes |
| 2 | CBX8 | 312 | 2.159 | 0.1962 | Yes |
| 3 | TNKS1BP1 | 543 | 1.886 | 0.2664 | Yes |
| 4 | ZNF618 | 704 | 1.735 | 0.3340 | Yes |
| 5 | NOP53 | 1023 | 1.481 | 0.3815 | Yes |
| 6 | EZH1 | 1117 | 1.418 | 0.4388 | Yes |
| 7 | HSF1 | 1237 | 1.351 | 0.4917 | Yes |
| 8 | SALL1 | 1682 | 1.106 | 0.5156 | Yes |
| 9 | CHD5 | 1783 | 1.059 | 0.5567 | Yes |
| 10 | DNMT3A | 1804 | 1.052 | 0.6019 | Yes |
| 11 | POLE3 | 1848 | 1.030 | 0.6449 | Yes |
| 12 | PHC2 | 2815 | 0.657 | 0.6200 | Yes |
| 13 | EME1 | 3172 | 0.572 | 0.6253 | Yes |
| 14 | NCAPD3 | 3328 | 0.538 | 0.6404 | Yes |
| 15 | DNMT3L | 3362 | 0.530 | 0.6619 | Yes |
| 16 | SIRT6 | 3441 | 0.516 | 0.6802 | Yes |
| 17 | TASOR | 5692 | 0.249 | 0.5657 | No |
| 18 | RRP8 | 5889 | 0.237 | 0.5652 | No |
| 19 | MPHOSPH8 | 7116 | 0.178 | 0.5047 | No |
| 20 | RRP1B | 7161 | 0.176 | 0.5100 | No |
| 21 | TOP2B | 7727 | 0.156 | 0.4853 | No |
| 22 | CENPC | 8393 | 0.135 | 0.4542 | No |
| 23 | SUZ12 | 8607 | 0.129 | 0.4480 | No |
| 24 | LRWD1 | 8724 | 0.126 | 0.4470 | No |
| 25 | MBD2 | 9150 | 0.115 | 0.4284 | No |
| 26 | ESCO2 | 9796 | 0.101 | 0.3969 | No |
| 27 | BAZ1B | 9998 | 0.097 | 0.3899 | No |
| 28 | MECP2 | 10183 | 0.094 | 0.3838 | No |
| 29 | WDR76 | 10447 | 0.089 | 0.3731 | No |
| 30 | BAZ1A | 10532 | 0.088 | 0.3722 | No |
| 31 | MORC2 | 11841 | 0.066 | 0.3022 | No |
| 32 | HELLS | 12007 | 0.064 | 0.2958 | No |
| 33 | SIRT1 | 12011 | 0.064 | 0.2984 | No |
| 34 | ANAPC7 | 12017 | 0.063 | 0.3009 | No |
| 35 | SMARCA5 | 12172 | 0.062 | 0.2951 | No |
| 36 | KDM4C | 12363 | 0.059 | 0.2871 | No |
| 37 | KDM4A | 12453 | 0.058 | 0.2846 | No |
| 38 | KMT5C | 12524 | 0.057 | 0.2832 | No |
| 39 | HDAC1 | 12722 | 0.054 | 0.2746 | No |
| 40 | BAZ2A | 12734 | 0.054 | 0.2764 | No |
| 41 | SMARCAD1 | 12755 | 0.054 | 0.2776 | No |
| 42 | ORC2 | 12943 | 0.051 | 0.2694 | No |
| 43 | BEND3 | 12995 | 0.051 | 0.2688 | No |
| 44 | PCGF2 | 13162 | 0.048 | 0.2617 | No |
| 45 | PHC1 | 13779 | 0.040 | 0.2291 | No |
| 46 | ZBTB18 | 14263 | 0.030 | 0.2035 | No |
| 47 | KDM4D | 14285 | 0.029 | 0.2036 | No |
| 48 | H2AZ1 | 14333 | 0.027 | 0.2022 | No |
| 49 | SUV39H1 | 14402 | 0.022 | 0.1994 | No |
| 50 | MBD3 | 14732 | -0.000 | 0.1810 | No |
| 51 | ATRX | 14811 | -0.000 | 0.1767 | No |
| 52 | SALL4 | 14937 | -0.000 | 0.1697 | No |
| 53 | CHRAC1 | 15042 | -0.000 | 0.1639 | No |
| 54 | EZH2 | 15104 | -0.000 | 0.1605 | No |
| 55 | CBX1 | 15153 | -0.000 | 0.1578 | No |
| 56 | MACROH2A1 | 15280 | -0.000 | 0.1508 | No |
| 57 | CENPA | 15308 | -0.000 | 0.1493 | No |
| 58 | TCP1 | 15438 | -0.000 | 0.1421 | No |
| 59 | RNF2 | 15467 | -0.000 | 0.1405 | No |
| 60 | CBX3 | 15487 | -0.000 | 0.1394 | No |
| 61 | SNAI1 | 15536 | -0.000 | 0.1368 | No |
| 62 | CENPB | 15578 | -0.000 | 0.1345 | No |
| 63 | TRIM28 | 15689 | -0.000 | 0.1283 | No |
| 64 | DNMT1 | 15690 | -0.000 | 0.1283 | No |
| 65 | UBA1 | 15694 | -0.000 | 0.1282 | No |
| 66 | UHRF2 | 16136 | -0.000 | 0.1036 | No |
| 67 | CDKN2A | 16138 | -0.000 | 0.1035 | No |
| 68 | HMGA2 | 16168 | -0.000 | 0.1019 | No |
| 69 | BMI1 | 16687 | -0.000 | 0.0730 | No |
| 70 | H1-4 | 16688 | -0.000 | 0.0730 | No |
| 71 | CBX2 | 16931 | -0.000 | 0.0595 | No |
| 72 | CBX6 | 17206 | -0.000 | 0.0442 | No |
| 73 | H1-5 | 17229 | -0.000 | 0.0430 | No |
| 74 | IKZF1 | 17289 | -0.000 | 0.0397 | No |
| 75 | RING1 | 17616 | -0.000 | 0.0215 | No |
| 76 | UHRF1 | 17954 | -0.000 | 0.0027 | No |